MicroRNA-resistant alleles of HOMEOBOX DOMAIN-2 modify inflorescence branching and increase grain protein content of wheat
- PMID: 35544571
- PMCID: PMC9094671
- DOI: 10.1126/sciadv.abn5907
MicroRNA-resistant alleles of HOMEOBOX DOMAIN-2 modify inflorescence branching and increase grain protein content of wheat
Abstract
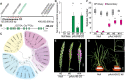
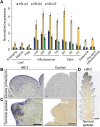
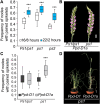
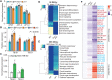
Plant and inflorescence architecture determine the yield potential of crops. Breeders have harnessed natural diversity for inflorescence architecture to improve yields, and induced genetic variation could provide further gains. Wheat is a vital source of protein and calories; however, little is known about the genes that regulate the development of its inflorescence. Here, we report the identification of semidominant alleles for a class III homeodomain-leucine zipper transcription factor, HOMEOBOX DOMAIN-2 (HB-2), on wheat A and D subgenomes, which generate more flower-bearing spikelets and enhance grain protein content. These alleles increase HB-2 expression by disrupting a microRNA 165/166 complementary site with conserved roles in plants; higher HB-2 expression is associated with modified leaf and vascular development and increased amino acid supply to the inflorescence during grain development. These findings enhance our understanding of genes that control wheat inflorescence development and introduce an approach to improve the nutritional quality of grain.
Figures



References
-
- Shiferaw B., Smale M., Braun H. J., Duveiller E., Reynolds M., Muricho G., Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security 5, 291–317 (2013).
-
- Reynolds M., Foulkes M. J., Slafer G. A., Berry P., Parry M. A. J., Snape J. W., Angus W. J., Raising yield potential in wheat. J. Exp. Bot. 60, 1899–1918 (2009). - PubMed
-
- Sreenivasulu N., Schnurbusch T., A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 17, 91–101 (2012). - PubMed
-
- Adamski N. M., Borrill P., Brinton J., Harrington S. A., Marchal C., Bentley A. R., Bovill W. D., Cattivelli L., Cockram J., C.-Moreira B., Ford B., Ghosh S., Harwood W., H.-Pak K., Hayta S., Hickey L. T., Kanyuka K., King J., Maccaferrri M., Naamati G., Pozniak C. J., R.-Gonzalez R. H., Sansaloni C., Trevaskis B., Wingen L. U., Wulff B. B. H., Uauy C., A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat. eLife 9, e55646 (2020). - PMC - PubMed
-
- IWGSC , Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7161 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

