Therapeutic targeting of LCK tyrosine kinase and mTOR signaling in T-cell acute lymphoblastic leukemia
- PMID: 35544598
- PMCID: PMC10082361
- DOI: 10.1182/blood.2021015106
Therapeutic targeting of LCK tyrosine kinase and mTOR signaling in T-cell acute lymphoblastic leukemia
Abstract
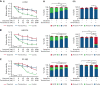
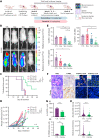
Relapse and refractory T-cell acute lymphoblastic leukemia (T-ALL) has a poor prognosis, and new combination therapies are sorely needed. Here, we used an ex vivo high-throughput screening platform to identify drug combinations that kill zebrafish T-ALL and then validated top drug combinations for preclinical efficacy in human disease. This work uncovered potent drug synergies between AKT/mTORC1 (mammalian target of rapamycin complex 1) inhibitors and the general tyrosine kinase inhibitor dasatinib. Importantly, these same drug combinations effectively killed a subset of relapse and dexamethasone-resistant zebrafish T-ALL. Clinical trials are currently underway using the combination of mTORC1 inhibitor temsirolimus and dasatinib in other pediatric cancer indications, leading us to prioritize this therapy for preclinical testing. This combination effectively curbed T-ALL growth in human cell lines and primary human T-ALL and was well tolerated and effective in suppressing leukemia growth in patient-derived xenografts (PDX) grown in mice. Mechanistically, dasatinib inhibited phosphorylation and activation of the lymphocyte-specific protein tyrosine kinase (LCK) to blunt the T-cell receptor (TCR) signaling pathway, and when complexed with mTORC1 inhibition, induced potent T-ALL cell killing through reducing MCL-1 protein expression. In total, our work uncovered unexpected roles for the LCK kinase and its regulation of downstream TCR signaling in suppressing apoptosis and driving continued leukemia growth. Analysis of a wide array of primary human T-ALLs and PDXs grown in mice suggest that combination of temsirolimus and dasatinib treatment will be efficacious for a large fraction of human T-ALLs.
© 2022 by The American Society of Hematology.
Conflict of interest statement
Figures







References
-
- Tremblay CS, Hoang T, Hoang T. Early T cell differentiation lessons from T-cell acute lymphoblastic leukemia. Prog Mol Biol Transl Sci. 2010;92(C):121–156. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

