CancerVar: An artificial intelligence-empowered platform for clinical interpretation of somatic mutations in cancer
- PMID: 35544644
- PMCID: PMC9075800
- DOI: 10.1126/sciadv.abj1624
CancerVar: An artificial intelligence-empowered platform for clinical interpretation of somatic mutations in cancer
Abstract
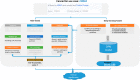
Several knowledgebases are manually curated to support clinical interpretations of thousands of hotspot somatic mutations in cancer. However, discrepancies or even conflicting interpretations are observed among these databases. Furthermore, many previously undocumented mutations may have clinical or functional impacts on cancer but are not systematically interpreted by existing knowledgebases. To address these challenges, we developed CancerVar to facilitate automated and standardized interpretations for 13 million somatic mutations based on the AMP/ASCO/CAP 2017 guidelines. We further introduced a deep learning framework to predict oncogenicity for these variants using both functional and clinical features. CancerVar achieved satisfactory performance when compared to several independent knowledgebases and, using clinically curated datasets, demonstrated practical utility in classifying somatic variants. In summary, by integrating clinical guidelines with a deep learning framework, CancerVar facilitates clinical interpretation of somatic variants, reduces manual work, improves consistency in variant classification, and promotes implementation of the guidelines.
Figures

References
-
- Chakravarty D., Gao J., Phillips S. M., Kundra R., Zhang H., Wang J., Rudolph J. E., Yaeger R., Soumerai T., Nissan M. H., Chang M. T., Chandarlapaty S., Traina T. A., Paik P. K., Ho A. L., Hantash F. M., Grupe A., Baxi S. S., Callahan M. K., Snyder A., Chi P., Danila D., Gounder M., Harding J. J., Hellmann M. D., Iyer G., Janjigian Y., Kaley T., Levine D. A., Lowery M., Omuro A., Postow M. A., Rathkopf D., Shoushtari A. N., Shukla N., Voss M., Paraiso E., Zehir A., Berger M. F., Taylor B. S., Saltz L. B., Riely G. J., Ladanyi M., Hyman D. M., Baselga J., Sabbatini P., Solit D. B., Schultz N., OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 2017, (2017). - PMC - PubMed
-
- Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Ng P. K.-S., Jeong K. J., Cao S., Wang Z., Gao J., Gao Q., Wang F., Liu E. M., Mularoni L., Rubio-Perez C., Nagarajan N., Cortes-Ciriano I., Zhou D. C., Liang W. W., Hess J. M., Yellapantula V. D., Tamborero D., Gonzalez-Perez A., Suphavilai C., Ko J. Y., Khurana E., Park P. J., Van Allen E. M., Liang H.; MC3 Working Group; Cancer Genome Atlas Research Network, Lawrence M. S., Lawrence M. S., Godzik A., Lopez-Bigas N., Stuart J., Wheeler D., Getz G., Chen K., Lazar A. J., Mills G. B., Karchin R., Ding L., Comprehensive characterization of cancer driver genes and mutations. Cell 174, 1034–1035 (2018). - PMC - PubMed
-
- Micheel C. M., Sweeney S. M., LeNoue-Newton M. L., Andre F., Bedard P. L., Guinney J., Meijer G. A., Rollins B. J., Sawyers C. L., Schultz N., Shaw K. R. M., Velculescu V. E., Levy M. A.; AACR Project GENIE Consortium , American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange: From inception to first data release and beyond-lessons learned and member institutions’ perspectives. JCO Clin. Cancer Inform. 2, 1–14 (2018). - PMC - PubMed
-
- Griffith M., Spies N. C., Krysiak K., McMichael J. F., Coffman A. C., Danos A. M., Ainscough B. J., Ramirez C. A., Rieke D. T., Kujan L., Barnell E. K., Wagner A. H., Skidmore Z. L., Wollam A., Liu C. J., Jones M. R., Bilski R. L., Lesurf R., Feng Y. Y., Shah N. M., Bonakdar M., Trani L., Matlock M., Ramu A., Campbell K. M., Spies G. C., Graubert A. P., Gangavarapu K., Eldred J. M., Larson D. E., Walker J. R., Good B. M., Wu C., Su A. I., Dienstmann R., Margolin A. A., Tamborero D., Lopez-Bigas N., Jones S. J., Bose R., Spencer D. H., Wartman L. D., Wilson R. K., Mardis E. R., Griffith O. L., CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017). - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

