A proposed unified mitotic chromosome architecture
- PMID: 35544689
- PMCID: PMC9171755
- DOI: 10.1073/pnas.2119107119
A proposed unified mitotic chromosome architecture
Abstract
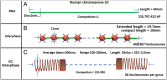
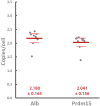
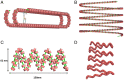
A molecular architecture is proposed for a representative mitotic chromosome, human chromosome 10. This architecture is built on an interphase chromosome structure based on cryo-electron microscopy (cryo-EM) cellular tomography [J. Sedat et al., Proc. Natl. Acad. Sci. U.S.A., in press], thus unifying chromosome structure throughout the complete mitotic cycle. The basic organizational principle for mitotic chromosomes is specific coiling of the 11-nm nucleosome fiber into large scale, ∼200-nm interphase structures, a Slinky [https://en.wikipedia.org/wiki/Slinky; motif cited in S. Bowerman et al., eLife 10, e65587 (2021)], then further modified with subsequent additional coiling for the final mitotic chromosome structure. The final mitotic chromosome architecture accounts for the dimensional values as well as the well-known cytological configurations. In addition, proof is experimentally provided by digital PCR technology that G1 T cell nuclei are diploid with one DNA molecule per chromosome. Many nucleosome linker DNA sequences, the promotors and enhancers, are suggestive of optimal exposure on the surfaces of the large-scale coils.
Keywords: biophysics and computational structure analysis; cell biology; chromosome structure; computational structure analysis.
Conflict of interest statement
The authors declare no competing interest.
Figures


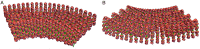

Similar articles
-
Helical coiled nucleosome chromosome architectures during cell cycle progression.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2410584121. doi: 10.1073/pnas.2410584121. Epub 2024 Oct 14. Proc Natl Acad Sci U S A. 2024. PMID: 39401359 Free PMC article.
-
A proposed unified interphase nucleus chromosome structure: Preliminary preponderance of evidence.Proc Natl Acad Sci U S A. 2022 Jun 28;119(26):e2119101119. doi: 10.1073/pnas.2119101119. Epub 2022 Jun 24. Proc Natl Acad Sci U S A. 2022. PMID: 35749363 Free PMC article.
-
New insight into the mitotic chromosome structure: irregular folding of nucleosome fibers without 30-nm chromatin structure.Cold Spring Harb Symp Quant Biol. 2010;75:439-44. doi: 10.1101/sqb.2010.75.034. Epub 2011 Mar 29. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21447821
-
Mitotic chromosome condensation in vertebrates.Exp Cell Res. 2012 Jul 15;318(12):1435-41. doi: 10.1016/j.yexcr.2012.03.017. Epub 2012 Mar 27. Exp Cell Res. 2012. PMID: 22475678 Review.
-
Genome Organization and Chromosome Architecture.Cold Spring Harb Symp Quant Biol. 2015;80:83-91. doi: 10.1101/sqb.2015.80.027318. Epub 2016 Jan 22. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26801160 Review.
Cited by
-
Helical coiled nucleosome chromosome architectures during cell cycle progression.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2410584121. doi: 10.1073/pnas.2410584121. Epub 2024 Oct 14. Proc Natl Acad Sci U S A. 2024. PMID: 39401359 Free PMC article.
-
Multi-Scale Imaging of the Dynamic Organization of Chromatin.Int J Mol Sci. 2023 Nov 4;24(21):15975. doi: 10.3390/ijms242115975. Int J Mol Sci. 2023. PMID: 37958958 Free PMC article. Review.
-
A proposed unified interphase nucleus chromosome structure: Preliminary preponderance of evidence.Proc Natl Acad Sci U S A. 2022 Jun 28;119(26):e2119101119. doi: 10.1073/pnas.2119101119. Epub 2022 Jun 24. Proc Natl Acad Sci U S A. 2022. PMID: 35749363 Free PMC article.
-
Nanoscale analysis of human G1 and metaphase chromatin in situ.EMBO J. 2025 May;44(9):2658-2694. doi: 10.1038/s44318-025-00407-2. Epub 2025 Mar 17. EMBO J. 2025. PMID: 40097852 Free PMC article.
-
Direct observation of surface charge and stiffness of human metaphase chromosomes.Nanoscale Adv. 2022 Dec 20;5(2):368-377. doi: 10.1039/d2na00620k. eCollection 2023 Jan 18. Nanoscale Adv. 2022. PMID: 36756276 Free PMC article.
References
-
- Sedat J., et al. , A proposed unified interphase nucleus chromosome structure: Preliminary preponderance of evidence. Proc. Natl. Acad. Sci. U.S.A. https://doi.org/10.1101/2021.10.08.463051. - PMC - PubMed
-
- Wikipedia, Slinky. https://en.wikipedia.org/wiki/Slinky. Accessed 28 April 2022.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

