Functional genome-wide short hairpin RNA library screening identifies key molecules for extracellular vesicle secretion from microglia
- PMID: 35545052
- PMCID: PMC9133589
- DOI: 10.1016/j.celrep.2022.110791
Functional genome-wide short hairpin RNA library screening identifies key molecules for extracellular vesicle secretion from microglia
Abstract
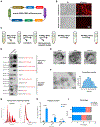
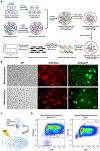
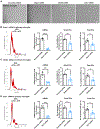
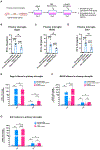
Activated microglia release extracellular vesicles (EVs) as modulators of brain homeostasis and innate immunity. However, the molecules critical for regulating EV production from microglia are poorly understood. Here we establish a murine microglial cell model to monitor EV secretion by measuring the fluorescence signal of tdTomato, which is linked to tetraspanin CD63. Stimulation of tdTomato+ cells with ATP induces rapid secretion of EVs and a reduction in cellular tdTomato intensity, reflecting EV secretion. We generate a GFP+ tdTomato+ cell library expressing TurboGFP and barcoded short hairpin RNAs for genome-wide screening using next-generation sequencing. We identify Mcfd2, Sepp1, and Sdc1 as critical regulators of ATP-induced EV secretion from murine microglia. Small interfering RNA (siRNA-based) silencing of each of these genes suppresses lipopolysaccharide- and ATP-induced inflammasome activation, as determined by interleukin-1β release from primary cultured murine microglia. These molecules are critical for microglial EV secretion and are potential therapeutic targets for neuroinflammatory disorders.
Keywords: CP: Neuroscience; extracellular vesicle; interleukin-1β; microglia; microtubule-associated protein tau; neuroinflammation; shRNA library screening.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declarations of interests T.I. had a sponsored research agreement with Ono Pharmaceutical and consults with Takeda.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

