Automated annotation of human centromeres with HORmon
- PMID: 35545449
- PMCID: PMC9248890
- DOI: 10.1101/gr.276362.121
Automated annotation of human centromeres with HORmon
Abstract
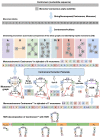
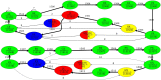
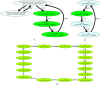
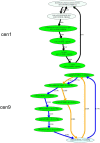
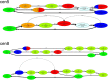
Recent advances in long-read sequencing opened a possibility to address the long-standing questions about the architecture and evolution of human centromeres. They also emphasized the need for centromere annotation (partitioning human centromeres into monomers and higher-order repeats [HORs]). Although there was a half-century-long series of semi-manual studies of centromere architecture, a rigorous centromere annotation algorithm is still lacking. Moreover, an automated centromere annotation is a prerequisite for studies of genetic diseases associated with centromeres and evolutionary studies of centromeres across multiple species. Although the monomer decomposition (transforming a centromere into a monocentromere written in the monomer alphabet) and the HOR decomposition (representing a monocentromere in the alphabet of HORs) are currently viewed as two separate problems, we show that they should be integrated into a single framework in such a way that HOR (monomer) inference affects monomer (HOR) inference. We thus developed the HORmon algorithm that integrates the monomer/HOR inference and automatically generates the human monomers/HORs that are largely consistent with the previous semi-manual inference.
© 2022 Kunyavskaya et al.; Published by Cold Spring Harbor Laboratory Press.
Figures








Similar articles
-
Novel Concept of Alpha Satellite Cascading Higher-Order Repeats (HORs) and Precise Identification of 15mer and 20mer Cascading HORs in Complete T2T-CHM13 Assembly of Human Chromosome 15.Int J Mol Sci. 2024 Apr 16;25(8):4395. doi: 10.3390/ijms25084395. Int J Mol Sci. 2024. PMID: 38673983 Free PMC article.
-
CentromereArchitect: inference and analysis of the architecture of centromeres.Bioinformatics. 2021 Jul 12;37(Suppl_1):i196-i204. doi: 10.1093/bioinformatics/btab265. Bioinformatics. 2021. PMID: 34252949 Free PMC article.
-
The string decomposition problem and its applications to centromere analysis and assembly.Bioinformatics. 2020 Jul 1;36(Suppl_1):i93-i101. doi: 10.1093/bioinformatics/btaa454. Bioinformatics. 2020. PMID: 32657390 Free PMC article.
-
Key-string algorithm--novel approach to computational analysis of repetitive sequences in human centromeric DNA.Croat Med J. 2003 Aug;44(4):386-406. Croat Med J. 2003. PMID: 12950141 Review.
-
What makes a centromere?Exp Cell Res. 2020 Apr 15;389(2):111895. doi: 10.1016/j.yexcr.2020.111895. Epub 2020 Feb 6. Exp Cell Res. 2020. PMID: 32035948 Review.
Cited by
-
De novo reconstruction of satellite repeat units from sequence data.ArXiv [Preprint]. 2023 Apr 19:arXiv:2304.09729v1. ArXiv. 2023. Update in: Genome Res. 2023 Dec 1;33(11):1994-2001. doi: 10.1101/gr.278005.123. PMID: 37131874 Free PMC article. Updated. Preprint.
-
Novel Cascade Alpha Satellite HORs in Orangutan Chromosome 13 Assembly: Discovery of the 59mer HOR-The largest Unit in Primates-And the Missing Triplet 45/27/18 HOR in Human T2T-CHM13v2.0 Assembly.Int J Mol Sci. 2024 Jul 11;25(14):7596. doi: 10.3390/ijms25147596. Int J Mol Sci. 2024. PMID: 39062839 Free PMC article.
-
Novel Concept of Alpha Satellite Cascading Higher-Order Repeats (HORs) and Precise Identification of 15mer and 20mer Cascading HORs in Complete T2T-CHM13 Assembly of Human Chromosome 15.Int J Mol Sci. 2024 Apr 16;25(8):4395. doi: 10.3390/ijms25084395. Int J Mol Sci. 2024. PMID: 38673983 Free PMC article.
-
Envisioning a new era: Complete genetic information from routine, telomere-to-telomere genomes.Am J Hum Genet. 2023 Nov 2;110(11):1832-1840. doi: 10.1016/j.ajhg.2023.09.011. Am J Hum Genet. 2023. PMID: 37922882 Free PMC article. Review.
-
The Satellite DNA PcH-Sat, Isolated and Characterized in the Limpet Patella caerulea (Mollusca, Gastropoda), Suggests the Origin from a Nin-SINE Transposable Element.Genes (Basel). 2024 Apr 25;15(5):541. doi: 10.3390/genes15050541. Genes (Basel). 2024. PMID: 38790169 Free PMC article.
References
-
- Ahuja RK, Magnati TL, Orlin JB. 1993. Network flows: theory, algorithms, and applications. Prentice-Hall, Upper Saddle River, NJ.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
