Comprehensive analysis of histone deacetylases genes in the prognosis and immune infiltration of glioma patients
- PMID: 35545840
- PMCID: PMC9134955
- DOI: 10.18632/aging.204071
Comprehensive analysis of histone deacetylases genes in the prognosis and immune infiltration of glioma patients
Abstract
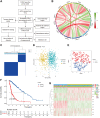
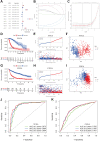
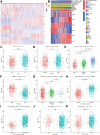
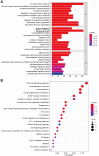
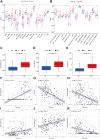
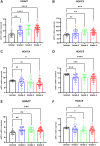
The occurrence and development of tumors are closely related to histone deacetylases (HDACs). However, their relationship with the overall biology and prognosis of glioma is still unknown. In the present study, we developed and validated a prognostic model for glioma based on HDAC genes. Glioma patients can be divided into two subclasses based on eleven HDAC genes, and patients from the two subclasses had markedly different survival outcomes. Then, using six HDAC genes (HDAC1, HDAC3, HDAC4, HDAC5, HDAC7, and HDAC9), we established a prognostic model for glioma patients, and this prognostic model was validated in an independent cohort. Furthermore, the calculated risk score from six HDACA genes expression was found to be an independent prognostic factor that could predict the five-year overall survival of glioma patients well. High-risk patients have changes in multiple complex functions and molecular signaling pathways, and the gene alterations of high- and low-risk patients were significantly different. We also found that the different survival outcomes of high- and low-risk patients could be related to the differences in immune filtration levels and the tumor microenvironment. Subsequently, we identified several small molecular compounds that could be favorable for glioma patient treatment. Finally, the expression levels of HDAC genes from the prognostic model were validated in glioma and nontumor tissue samples. Our results revealed the clinical utility and potential molecular mechanisms of HDAC genes in glioma. A model based on six HDAC genes can predict the overall survival of glioma patients well, and these genes are potential therapeutic targets.
Keywords: bioinformatics; glioma; histone deacetylases; immune infiltration; prognostic factor.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

