Pseudomonas aeruginosa Alters Peptidoglycan Composition under Nutrient Conditions Resembling Cystic Fibrosis Lung Infections
- PMID: 35545925
- PMCID: PMC9239049
- DOI: 10.1128/msystems.00156-22
Pseudomonas aeruginosa Alters Peptidoglycan Composition under Nutrient Conditions Resembling Cystic Fibrosis Lung Infections
Abstract
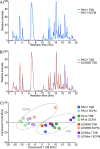
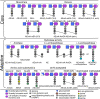
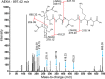
Epidemic strains of Pseudomonas aeruginosa are highly virulent opportunistic pathogens with increased transmissibility and enhanced antimicrobial resistance. Understanding the cellular mechanisms behind this heightened virulence and resistance is critical. Peptidoglycan (PG) is an integral component of P. aeruginosa cells that is essential to its survival and a target for antimicrobials. Here, we examined the global PG composition of two P. aeruginosa epidemic strains, LESB58 and LESlike1, and compared them to the common laboratory strains PAO1 and PA14. We also examined changes in PG composition when the strains were cultured under nutrient conditions that resembled cystic fibrosis lung infections. We identified 448 unique muropeptides and provide the first evidence for stem peptides modified with O-methylation, meso-diaminopimelic acid (mDAP) deamination, and novel substitutions of mDAP residues within P. aeruginosa PG. Our results also present the first evidence for both d,l- and l,d-endopeptidase activity on the PG sacculus of a Gram-negative organism. The PG composition of the epidemic strains varied significantly when grown under conditions resembling cystic fibrosis (CF) lung infections, showing increases in O-methylated stem peptides and decreases in l,d-endopeptidase activity as well as an increased abundance of de-N-acetylated sugars and l,d-transpeptidase activity, which are related to bacterial virulence and antibiotic resistance, respectively. We also identified strain-specific changes where LESlike1 increased the addition of unique amino acids to the terminus of the stem peptide and LESB58 increased amidase activity. Overall, this study demonstrates that P. aeruginosa PG composition is primarily influenced by nutrient conditions that mimic the CF lung; however, inherent strain-to-strain differences also exist. IMPORTANCE Using peptidoglycomics to examine the global composition of the peptidoglycan (PG) allows insights into the enzymatic activity that functions on this important biopolymer. Changes within the PG structure have implications for numerous physiological processes, including virulence and antimicrobial resistance. The identification of highly unique PG modifications illustrates the complexity of this biopolymer in Pseudomonas aeruginosa. Analyzing the PG composition of clinical P. aeruginosa epidemic strains provides insights into the increased virulence and antimicrobial resistance of these difficult-to-eradicate infections.
Keywords: Liverpool epidemic strain; Pseudomonas aeruginosa; antibiotic resistance; peptidoglycan; peptidoglycomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Cystic Fibrosis Foundation. 2020. Cystic fibrosis foundation patient registry 2019 annual data report. Cystic Fibrosis Foundation, Bethesda, MD.
-
- Aaron SD, Vandemheen KL, Ramotar K, Giesbrecht-Lewis T, Tullis E, Freitag A, Paterson N, Jackson M, Lougheed MD, Dowson C, Kumar V, Ferris W, Chan F, Doucette S, Fergusson D. 2010. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA 304:2145–2153. doi:10.1001/jama.2010.1665. - DOI - PubMed
-
- Parkins MD, Glezerson BA, Sibley CD, Sibley KA, Duong J, Purighalla S, Mody CH, Workentine ML, Storey DG, Surette MG, Rabin HR. 2014. Twenty-five-year outbreak of Pseudomonas aeruginosa infecting individuals with cystic fibrosis: identification of the prairie epidemic strain. J Clin Microbiol 52:1127–1135. doi:10.1128/JCM.03218-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

