Meta-analysis of sub-Saharan African studies provides insights into genetic architecture of lipid traits
- PMID: 35546142
- PMCID: PMC9095599
- DOI: 10.1038/s41467-022-30098-w
Meta-analysis of sub-Saharan African studies provides insights into genetic architecture of lipid traits
Erratum in
-
Author Correction: Meta-analysis of sub-Saharan African studies provides insights into genetic architecture of lipid traits.Nat Commun. 2022 Aug 2;13(1):4474. doi: 10.1038/s41467-022-32072-y. Nat Commun. 2022. PMID: 35918347 Free PMC article. No abstract available.
Abstract
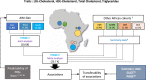
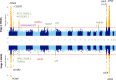
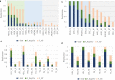
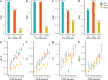
Genetic associations for lipid traits have identified hundreds of variants with clear differences across European, Asian and African studies. Based on a sub-Saharan-African GWAS for lipid traits in the population cross-sectional AWI-Gen cohort (N = 10,603) we report a novel LDL-C association in the GATB region (P-value=1.56 × 10-8). Meta-analysis with four other African cohorts (N = 23,718) provides supporting evidence for the LDL-C association with the GATB/FHIP1A region and identifies a novel triglyceride association signal close to the FHIT gene (P-value =2.66 × 10-8). Our data enable fine-mapping of several well-known lipid-trait loci including LDLR, PMFBP1 and LPA. The transferability of signals detected in two large global studies (GLGC and PAGE) consistently improves with an increase in the size of the African replication cohort. Polygenic risk score analysis shows increased predictive accuracy for LDL-C levels with the narrowing of genetic distance between the discovery dataset and our cohort. Novel discovery is enhanced with the inclusion of African data.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Blauw LL, et al. CETP (Cholesteryl Ester Transfer Protein) concentration: a genome-wide association study followed by mendelian randomization on coronary artery disease. Circ. Genom. Precis. Med. 2018;11:e002034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

