Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation
- PMID: 35546409
- PMCID: PMC9097414
- DOI: 10.1186/s40168-022-01272-5
Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation
Abstract
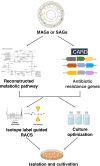
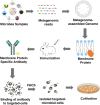
Although there is now an extensive understanding of the diversity of microbial life on earth through culture-independent metagenomic DNA sequence analyses, the isolation and cultivation of microbes remains critical to directly study them and confirm their metabolic and physiological functions, and their ecological roles. The majority of environmental microbes are as yet uncultured however; therefore, bringing these rare or poorly characterized groups into culture is a priority to further understand microbiome functions. Moreover, cultivated isolates may find utility in a range of applications, such as new probiotics, biocontrol agents, and agents for industrial processes. The growing abundance of metagenomic and meta-transcriptomic sequence information from a wide range of environments provides more opportunities to guide the isolation and cultivation of microbes of interest. In this paper, we discuss a range of successful methodologies and applications that have underpinned recent metagenome-guided isolation and cultivation of microbe efforts. These approaches include determining specific culture conditions to enrich for taxa of interest, to more complex strategies that specifically target the capture of microbial species through antibody engineering and genome editing strategies. With the greater degree of genomic information now available from uncultivated members, such as via metagenome-assembled genomes, the theoretical understanding of their cultivation requirements will enable greater possibilities to capture these and ultimately gain a more comprehensive understanding of the microbiomes. Video Abstract.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

