Patatin-like phospholipase CapV in Escherichia coli - morphological and physiological effects of one amino acid substitution
- PMID: 35546554
- PMCID: PMC9095652
- DOI: 10.1038/s41522-022-00294-z
Patatin-like phospholipase CapV in Escherichia coli - morphological and physiological effects of one amino acid substitution
Abstract
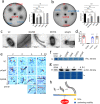
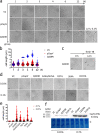
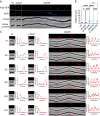
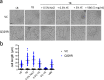
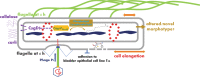
In rod-shaped bacteria, morphological plasticity occurs in response to stress, which blocks cell division to promote filamentation. We demonstrate here that overexpression of the patatin-like phospholipase variant CapVQ329R, but not CapV, causes pronounced sulA-independent pyridoxine-inhibited cell filamentation in the Escherichia coli K-12-derivative MG1655 associated with restriction of flagella production and swimming motility. Conserved amino acids in canonical patatin-like phospholipase A motifs, but not the nucleophilic serine, are required to mediate CapVQ329R phenotypes. Furthermore, CapVQ329R production substantially alters the lipidome and colony morphotype including rdar biofilm formation with modulation of the production of the biofilm activator CsgD, and affects additional bacterial traits such as the efficiency of phage infection and antimicrobial susceptibility. Moreover, genetically diverse commensal and pathogenic E. coli strains and Salmonella typhimurium responded with cell filamentation and modulation in colony morphotype formation to CapVQ329R expression. In conclusion, this work identifies the CapV variant CapVQ329R as a pleiotropic regulator, emphasizes a scaffold function for patatin-like phospholipases, and highlights the impact of the substitution of a single conserved amino acid for protein functionality and alteration of host physiology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

