HOXA10 Regulates the Synthesis of Cholesterol in Endometrial Stromal Cells
- PMID: 35546998
- PMCID: PMC9084188
- DOI: 10.3389/fendo.2022.852671
HOXA10 Regulates the Synthesis of Cholesterol in Endometrial Stromal Cells
Abstract
Background: The expression of homeobox A10 (HOXA10) in endometrial stromal cells is regulated by steroid hormones, especially by estrogen. As a precursor molecule of estrogen, abnormal cholesterol metabolism is significantly positively correlated with endometriosis. The purpose of this study was to explore the regulation of HOXA10 on cholesterol synthesis in endometrial stromal cells.
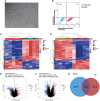
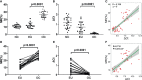
Method: mRNA expression data of eutopic endometrial stromal cell (ESC) and ovarian endometriotic cysts stromal cell (OESC) were download from the Gene Expression Omnibus (GEO) databases. Overexpression and silence of HOXA10 were conducted in cultured ESC and subjected to mRNA sequencing. The differentially expressed genes (DEGs) were selected by analyzing the sequencing data. Weighted gene co-expression network analysis (WGCNA) was applied to identify the key genes associated with HOXA10. The methylation rate of HOXA10 CpGs and the correlation between HOXA10 expression and the methylation in eutopic endometrial tissue (EU) and ovarian cyst (OC) were analyzed.
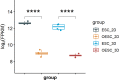
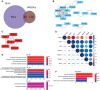
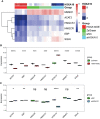
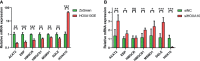
Results: HOXA10 in ESC was significantly higher expressed than that in OESC. Six key genes (HMGCR, MSMO1, ACAT2, HMGCS1, EBP, and SQLE), which were regulated by HOXA10, were identified from the salmon4 module by WGCNA. All these key genes were enriched in cholesterol synthesis. Moreover, the expression of HOXA10 was negatively related to its CpGs methylation rate.
Conclusion: In this study, six key genes that were regulated by HOXA10 were selected, and all of them were enriched in cholesterol synthesis. This finding provided a new insight into the metabolic mechanism of cholesterol in ESC. It also provided a potential treatment strategy for cholesterol metabolism maladjustment in patients with ovarian endometriosis.
Keywords: HOXA10 gene; cholesterol synthesis; endometrial stromal cell; estrogen; ovarian endometriosis.
Copyright © 2022 Yu, Tang, Huang, Guo, Du, Li and Quan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

