The Effects of Helicobacter pylori Infection on Gastric Microbiota in Children With Duodenal Ulcer
- PMID: 35547124
- PMCID: PMC9082302
- DOI: 10.3389/fmicb.2022.853184
The Effects of Helicobacter pylori Infection on Gastric Microbiota in Children With Duodenal Ulcer
Abstract
Background: Helicobacter pylori (H. pylori) infection is the main cause of chronic gastritis and duodenal ulcer in children. Little is known about the effect of H. pylori on gastric microbiota in children with duodenal ulcer. This study is aimed at the characteristics of gastric microbiota in children with duodenal ulcer on H. pylori infection.
Methods: We studied 23 children diagnosed with duodenal ulcer by gastric endoscopy because of the gastrointestinal symptoms, 15 children were diagnosed with H. pylori infection, while 8 children were without H. pylori infection. Endoscopic mucosal biopsy samples were obtained for DNA extraction. Microbiomes were analyzed by 16S rRNA profiling and microbial functions were predicted using the software Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt).
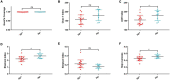
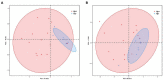
Results: Bacterial richness and diversity of gastric microbiota in duodenal ulcer with H. pylori-positive were lower than those negative. The gastric microbiota in H. pylori-positive group significantly reduced proportions of six phyla and fifteen genera; only Helicobacter taxa were more abundant in H. pylori-positive group. Co-expression network analysis showed a more complex network of interactions in the H. pylori-positive group than that in the H. pylori-negative group. For the predicted functions, lower abundance in the pathways of carbohydrate metabolism, signal transduction, amino acid metabolism, and lipid metabolism were found in H. pylori-positive group than the H. pylori-negative group. H. pylori colonization reduces a microbial community with genotoxic potential in the gastric mucosa of children with duodenal ulcer.
Conclusions: The presence of H. pylori significantly influences gastric microbiota and results in a lower abundance of multiple taxonomic levels in children with duodenal ulcer. Children with duodenal ulcer exhibit a dysbiotic microbial community with genotoxic potential, which is distinct from that of children with H. pylori infection.
Clinical trial registration: [http://www.chictr.org.cn], identifier [ChiCTR1800015190].
Keywords: 16S rRNA; Helicobacter pylori; children; duodenal ulcer; gastric microbiota.
Copyright © 2022 Zheng, Zhu, Ying, Long, Chen, Peng, Li, Zhao and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

