Extracting Dynamical Understanding From Neural-Mass Models of Mouse Cortex
- PMID: 35547660
- PMCID: PMC9081874
- DOI: 10.3389/fncom.2022.847336
Extracting Dynamical Understanding From Neural-Mass Models of Mouse Cortex
Abstract
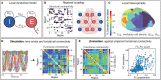
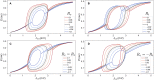
New brain atlases with high spatial resolution and whole-brain coverage have rapidly advanced our knowledge of the brain's neural architecture, including the systematic variation of excitatory and inhibitory cell densities across the mammalian cortex. But understanding how the brain's microscale physiology shapes brain dynamics at the macroscale has remained a challenge. While physiologically based mathematical models of brain dynamics are well placed to bridge this explanatory gap, their complexity can form a barrier to providing clear mechanistic interpretation of the dynamics they generate. In this work, we develop a neural-mass model of the mouse cortex and show how bifurcation diagrams, which capture local dynamical responses to inputs and their variation across brain regions, can be used to understand the resulting whole-brain dynamics. We show that strong fits to resting-state functional magnetic resonance imaging (fMRI) data can be found in surprisingly simple dynamical regimes-including where all brain regions are confined to a stable fixed point-in which regions are able to respond strongly to variations in their inputs, consistent with direct structural connections providing a strong constraint on functional connectivity in the anesthetized mouse. We also use bifurcation diagrams to show how perturbations to local excitatory and inhibitory coupling strengths across the cortex, constrained by cell-density data, provide spatially dependent constraints on resulting cortical activity, and support a greater diversity of coincident dynamical regimes. Our work illustrates methods for visualizing and interpreting model performance in terms of underlying dynamical mechanisms, an approach that is crucial for building explanatory and physiologically grounded models of the dynamical principles that underpin large-scale brain activity.
Keywords: brain dynamics; cell densities; dynamical systems; mouse cortex; neural mass model.
Copyright © 2022 Siu, Müller, Zerbi, Aquino and Fulcher.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Allegra Mascaro A. L., Falotico E., Petkoski S., Pasquini M., Vannucci L., Tort-Colet N., et al. (2020). Experimental and computational study on motor control and recovery after stroke: toward a constructive loop between experimental and virtual embodied neuroscience. Front. Syst. Neurosci. 14, 31. 10.3389/fnsys.2020.00031 - DOI - PMC - PubMed
-
- Borisyuk R. M., Kirillov A. B. (1992). Bifurcation analysis of a neural network model. Biol. Cybern. 66, 319–325. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

