Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron
- PMID: 35547740
- PMCID: PMC9082315
- DOI: 10.3389/fimmu.2022.863831
Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron
Abstract
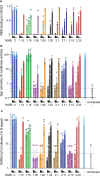
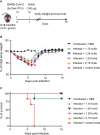
The emergence of SARS-CoV-2 variants that escape from immune neutralization are challenging vaccines and antibodies developed to stop the COVID-19 pandemic. Thus, it is important to establish therapeutics directed toward multiple or specific SARS-CoV-2 variants. The envelope spike (S) glycoprotein of SARS-CoV-2 is the key target of neutralizing antibodies (Abs). We selected a panel of nine nanobodies (Nbs) from dromedary camels immunized with the receptor-binding domain (RBD) of the S, and engineered Nb fusions as humanized heavy chain Abs (hcAbs). Nbs and derived hcAbs bound with subnanomolar or picomolar affinities to the S and its RBD, and S-binding cross-competition clustered them in two different groups. Most of the hcAbs hindered RBD binding to its human ACE2 (hACE2) receptor, blocked cell entry of viruses pseudotyped with the S protein and neutralized SARS-CoV-2 infection in cell cultures. Four potent neutralizing hcAbs prevented the progression to lethal SARS-CoV-2 infection in hACE2-transgenic mice, demonstrating their therapeutic potential. Cryo-electron microscopy identified Nb binding epitopes in and out the receptor binding motif (RBM), and showed different ways to prevent virus binding to its cell entry receptor. The Nb binding modes were consistent with its recognition of SARS-CoV-2 RBD variants; mono and bispecific hcAbs efficiently bound all variants of concern except omicron, which emphasized the immune escape capacity of this latest variant.
Keywords: COVID-19; SARS-CoV-2 variants; coronavirus; nanobodies; neutralizing antibodies.
Copyright © 2022 Casasnovas, Margolles, Noriega, Guzmán, Arranz, Melero, Casanova, Corbera, Jiménez-de-Oya, Gastaminza, Garaigorta, Saiz, Martín-Acebes and Fernández.
Conflict of interest statement
JMC, YM, MN, PG, UG, JS, MM-A, JAC, and LF are co-inventors on patent applications covering the Nb and hcAb molecules described in this manuscript. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

