Multi-scale integrative analyses identify THBS2+ cancer-associated fibroblasts as a key orchestrator promoting aggressiveness in early-stage lung adenocarcinoma
- PMID: 35547750
- PMCID: PMC9065207
- DOI: 10.7150/thno.69590
Multi-scale integrative analyses identify THBS2+ cancer-associated fibroblasts as a key orchestrator promoting aggressiveness in early-stage lung adenocarcinoma
Abstract
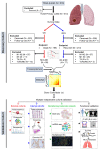
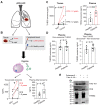
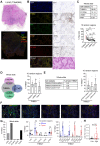
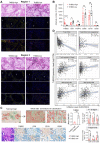
Rationale: Subsets of patients with early-stage lung adenocarcinoma (LUAD) have a poor post-surgical course after curative surgery. However, biomarkers stratifying this high-risk subset and molecular underpinnings underlying the aggressive phenotype remain unclear. Methods: We integrated bulk and single-cell transcriptomics, proteomics, secretome and spatial profiling of clinical early-stage LUAD samples to identify molecular underpinnings that promote the aggressive phenotype. Results: We identified and validated THBS2, at multi-omic levels, as a tumor size-independent biomarker that robustly predicted post-surgical survival in multiple independent clinical cohorts of early-stage LUAD. Furthermore, scRNA-seq data revealed that THBS2 is exclusively derived from a specific cancer-associated fibroblast (CAF) subset that is distinct from CAFs defined by classical markers. Interestingly, our data demonstrated that THBS2 was preferentially secreted via exosomes in early-stage LUAD tumors with high aggressiveness, and its levels in the peripheral plasma associated with short recurrence-free survival. Further characterization showed that THBS2-high early-stage LUAD was characterized by suppressed antitumor immunity. Specifically, beyond tumor cells, THBS2+ CAFs mainly interact with B and CD8+ T lymphocytes as well as macrophages within tumor microenvironment of early-stage LUAD, and THBS2-high LUAD was associated with decreased immune cell infiltrates but increased immune exhaustion marker. Clinically, high THBS2 expression predicted poor response to immunotherapies and short post-treatment survival of patients. Finally, THBS2 recombinant protein suppressed ex vivo T cells proliferation and promoted in vivo LUAD tumor growth and distant micro-metastasis. Conclusions: Our multi-level analyses uncovered tumor-specific THBS2+ CAFs as a key orchestrator promoting aggressiveness in early-stage LUAD.
Keywords: THBS2; cancer-associated fibroblast; early-stage lung adenocarcinoma; exosome; immunotherapy.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- de Koning HJ, van der Aalst CM, de Jong PA, Scholten ET, Nackaerts K, Heuvelmans MA. et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med. 2020;382:503–13. - PubMed
-
- Zhang J, Wu J, Tan Q, Zhu L, Gao W. Why do pathological stage IA lung adenocarcinomas vary from prognosis?: a clinicopathologic study of 176 patients with pathological stage IA lung adenocarcinoma based on the IASLC/ATS/ERS classification. J Thorac Oncol. 2013;8:1196–202. - PubMed
-
- Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG. et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–64. - PubMed
-
- Hung JJ, Yeh YC, Jeng WJ, Wu KJ, Huang BS, Wu YC. et al. Predictive value of the international association for the study of lung cancer/American Thoracic Society/European Respiratory Society classification of lung adenocarcinoma in tumor recurrence and patient survival. J Clin Oncol. 2014;32:2357–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

