This is a preprint.
Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin
- PMID: 35547846
- PMCID: PMC9094096
- DOI: 10.1101/2021.09.15.460543
Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin
Update in
-
Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.Nat Commun. 2023 Apr 25;14(1):2366. doi: 10.1038/s41467-023-38031-5. Nat Commun. 2023. PMID: 37185902 Free PMC article.
Abstract
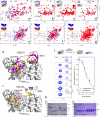
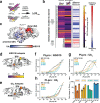
The Papain-like protease (PLpro) is a domain of a multi-functional, non-structural protein 3 of coronaviruses. PLpro cleaves viral polyproteins and posttranslational conjugates with poly-ubiquitin and protective ISG15, composed of two ubiquitin-like (UBL) domains. Across coronaviruses, PLpro showed divergent selectivity for recognition and cleavage of posttranslational conjugates despite sequence conservation. We show that SARS-CoV-2 PLpro binds human ISG15 and K48-linked di-ubiquitin (K48-Ub 2 ) with nanomolar affinity and detect alternate weaker-binding modes. Crystal structures of untethered PLpro complexes with ISG15 and K48-Ub 2 combined with solution NMR and cross-linking mass spectrometry revealed how the two domains of ISG15 or K48-Ub 2 are differently utilized in interactions with PLpro. Analysis of protein interface energetics predicted differential binding stabilities of the two UBL/Ub domains that were validated experimentally. We emphasize how substrate recognition can be tuned to cleave specifically ISG15 or K48-Ub 2 modifications while retaining capacity to cleave mono-Ub conjugates. These results highlight alternative druggable surfaces that would inhibit PLpro function.
Conflict of interest statement
COMPETING INTEREST
The authors declare that they have no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
