High-throughput lipidomics reveal mirabilite regulating lipid metabolism as anticancer therapeutics
- PMID: 35547938
- PMCID: PMC9087915
- DOI: 10.1039/c8ra06190d
High-throughput lipidomics reveal mirabilite regulating lipid metabolism as anticancer therapeutics
Abstract
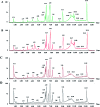
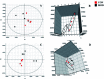
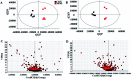
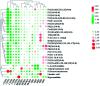
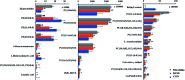
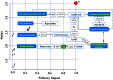
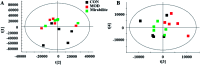
Altered lipid metabolism is an emerging hallmark of cancers. Mirabilite has a therapeutic effect on colorectal cancer (CRC); however, its metabolic mechanism remains unclear. This study aims to explore the potential therapeutic targets of mirabilite protection against colorectal cancer in APCmin/+ mice model. Oral administration of mirabilite was started from the ninth month, while the same dosage of distilled water was given to both the control group and the model group. Based on lipidomics, we collected serum samples of all mice at the 20th week and used a non-targeted method to identify the lipid biomarkers of CRC. Compared with C57BL/6J mice, the metabolic profile of CRC model mice was significantly disturbed, and we identified that 25 lipid-related biomarkers, including linoleic acid, 2-hydroxybutyric acid, 6-deoxocastasterone, hypoxanthine, PC(16:1), PC(18:4), and retinyl acetate, were associated with CRC. According to the abovementioned results, there were six lipid molecules with significant differences that can be used as new targets for handling of CRC through six metabolic pathways, namely, linoleic acid metabolism, retinol metabolism, propanoate metabolism, arachidonic acid metabolism, biosynthesis of unsaturated fatty acids and purine metabolism. Compared with the model group, the metabolic profiles of these disorders tend to recover after treatment. These results indicated that the lipid molecules associated with CRC were regulated by mirabilite. In addition, we identified seven key lipid molecules, of which four had statistical significance. After administration of mirabilite, all disordered metabolic pathways showed different degrees of regulation. In conclusion, high-throughput lipidomics approach revealed mirabilite regulating the altered lipid metabolism as anticancer therapeutics.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Ultra-performance liquid chromatography/mass spectrometry technology and high-throughput metabolomics for deciphering the preventive mechanism of mirabilite on colorectal cancer via the modulation of complex metabolic networks.RSC Adv. 2019 Oct 31;9(61):35356-35363. doi: 10.1039/c9ra07687e. eCollection 2019 Oct 31. RSC Adv. 2019. PMID: 35528071 Free PMC article.
-
High-throughput lipidomics characterize key lipid molecules as potential therapeutic targets of Kaixinsan protects against Alzheimer's disease in APP/PS1 transgenic mice.J Chromatogr B Analyt Technol Biomed Life Sci. 2018 Aug 15;1092:286-295. doi: 10.1016/j.jchromb.2018.06.032. Epub 2018 Jun 18. J Chromatogr B Analyt Technol Biomed Life Sci. 2018. PMID: 29933222
-
Network pharmacology combined with functional metabolomics discover bile acid metabolism as a promising target for mirabilite against colorectal cancer.RSC Adv. 2018 Aug 24;8(53):30061-30070. doi: 10.1039/c8ra04886j. eCollection 2018 Aug 24. RSC Adv. 2018. PMID: 35546810 Free PMC article.
-
Dysregulated lipid metabolism in colorectal cancer.Curr Opin Gastroenterol. 2022 Mar 1;38(2):162-167. doi: 10.1097/MOG.0000000000000811. Curr Opin Gastroenterol. 2022. PMID: 35098938 Review.
-
Targeting Lipid Metabolic Reprogramming as Anticancer Therapeutics.J Cancer Prev. 2016 Dec;21(4):209-215. doi: 10.15430/JCP.2016.21.4.209. Epub 2016 Dec 30. J Cancer Prev. 2016. PMID: 28053954 Free PMC article. Review.
Cited by
-
High-throughput metabolomics for evaluating the efficacy and discovering the metabolic mechanism of Luozhen capsules from the excessive liver-fire syndrome of hypertension.RSC Adv. 2019 Oct 9;9(55):32141-32153. doi: 10.1039/c9ra06622e. eCollection 2019 Oct 7. RSC Adv. 2019. PMID: 35530762 Free PMC article.
-
Chinmedomics: a potent tool for the evaluation of traditional Chinese medicine efficacy and identification of its active components.Chin Med. 2024 Mar 13;19(1):47. doi: 10.1186/s13020-024-00917-x. Chin Med. 2024. PMID: 38481256 Free PMC article. Review.
-
Untargeted metabolomics using liquid chromatography coupled with mass spectrometry for rapid discovery of metabolite biomarkers to reveal therapeutic effects of Psoralea corylifolia seeds against osteoporosis.RSC Adv. 2019 Nov 1;9(61):35429-35442. doi: 10.1039/c9ra07382e. eCollection 2019 Oct 31. RSC Adv. 2019. PMID: 35528068 Free PMC article.
-
Identification of key lipid metabolites during metabolic dysregulation in the diabetic retinopathy disease mouse model and efficacy of Keluoxin capsule using an UHPLC-MS-based non-targeted lipidomics approach.RSC Adv. 2021 Feb 1;11(10):5491-5505. doi: 10.1039/d0ra00343c. eCollection 2021 Jan 28. RSC Adv. 2021. PMID: 35423075 Free PMC article.
-
Mineral crude drug mirabilite (Mangxiao) inhibits the occurrence of colorectal cancer by regulating the Lactobacillus-bile acid-intestinal farnesoid X receptor axis based on multiomics integration analysis.MedComm (2020). 2024 Apr 25;5(5):e556. doi: 10.1002/mco2.556. eCollection 2024 May. MedComm (2020). 2024. PMID: 38665997 Free PMC article.
References
LinkOut - more resources
Full Text Sources

