The B1 Domain of Streptococcal Protein G Serves as a Multi-Functional Tag for Recombinant Protein Production in Plants
- PMID: 35548280
- PMCID: PMC9083265
- DOI: 10.3389/fpls.2022.878677
The B1 Domain of Streptococcal Protein G Serves as a Multi-Functional Tag for Recombinant Protein Production in Plants
Abstract
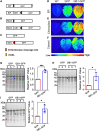
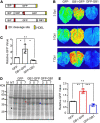
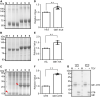
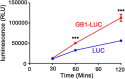
Plants have long been considered a cost-effective platform for recombinant production. A recently recognized additional advantage includes the low risk of contamination of human pathogens, such as viruses and bacterial endotoxins. Indeed, a great advance has been made in developing plants as a "factory" to produce recombinant proteins to use for biopharmaceutical purposes. However, there is still a need to develop new tools for recombinant protein production in plants. In this study, we provide data showing that the B1 domain of Streptococcal protein G (GB1) can be a multi-functional domain of recombinant proteins in plants. N-terminal fusion of the GB1 domain increased the expression level of various target proteins ranging from 1.3- to 3.1-fold at the protein level depending on the target proteins. GB1 fusion led to the stabilization of the fusion proteins. Furthermore, the direct detection of GB1-fusion proteins by the secondary anti-IgG antibody eliminated the use of the primary antibody for western blot analysis. Based on these data, we propose that the small GB1 domain can be used as a versatile tag for recombinant protein production in plants.
Keywords: GB1; Nicotiana benthamiana; biopharmaceutical proteins; plant-based molecular pharming; protein folding.
Copyright © 2022 Song, Diao, Moon, Yun and Hwang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
1H, 15N, and 13C chemical shift assignments of the micelle immersed FAT C-terminal (FATC) domains of the human protein kinases ataxia-telangiectasia mutated (ATM) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) fused to the B1 domain of streptococcal protein G (GB1).Biomol NMR Assign. 2018 Apr;12(1):149-154. doi: 10.1007/s12104-018-9798-3. Epub 2018 Jan 18. Biomol NMR Assign. 2018. PMID: 29349619
-
A fast and simple method for probing the interaction of peptides and proteins with lipids and membrane-mimetics using GB1 fusion proteins and NMR spectroscopy.Protein Sci. 2012 Oct;21(10):1566-70. doi: 10.1002/pro.2127. Protein Sci. 2012. PMID: 22825779 Free PMC article.
-
Enhancement of solubility and yield of a β-glucan receptor Dectin-1 C-type lectin-like domain in Escherichia coli with a solubility-enhancement tag.Protein Expr Purif. 2016 Jul;123:97-104. doi: 10.1016/j.pep.2016.04.002. Epub 2016 Apr 5. Protein Expr Purif. 2016. PMID: 27062941
-
Plant-made immunotoxin building blocks: A roadmap for producing therapeutic antibody-toxin fusions.Biotechnol Adv. 2021 Mar-Apr;47:107683. doi: 10.1016/j.biotechadv.2020.107683. Epub 2020 Dec 27. Biotechnol Adv. 2021. PMID: 33373687 Review.
-
Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production.Plants (Basel). 2020 Jul 4;9(7):842. doi: 10.3390/plants9070842. Plants (Basel). 2020. PMID: 32635427 Free PMC article. Review.
Cited by
-
Advances in Subcellular Accumulation Design for Recombinant Protein Production in Tobacco.Biodes Res. 2024 Aug 28;6:0047. doi: 10.34133/bdr.0047. eCollection 2024. Biodes Res. 2024. PMID: 39206181 Free PMC article. Review.
-
Production of active Exendin-4 in Nicotiana benthamiana and its application in treatment of type-2 diabetics.Front Plant Sci. 2022 Dec 22;13:1062658. doi: 10.3389/fpls.2022.1062658. eCollection 2022. Front Plant Sci. 2022. PMID: 36618620 Free PMC article.
-
SARS-CoV-2 spike trimer vaccine expressed in Nicotiana benthamiana adjuvanted with Alum elicits protective immune responses in mice.Plant Biotechnol J. 2022 Dec;20(12):2298-2312. doi: 10.1111/pbi.13908. Epub 2022 Sep 5. Plant Biotechnol J. 2022. PMID: 36062974 Free PMC article.
-
Production of active human iduronate-2-sulfatase (IDS) enzyme in Nicotiana benthamiana.Sci Rep. 2024 Oct 4;14(1):23066. doi: 10.1038/s41598-024-73778-x. Sci Rep. 2024. PMID: 39367006 Free PMC article.
-
Structure of saguaro cactus virus 3' translational enhancer mimics 5' cap for eIF4E binding.Proc Natl Acad Sci U S A. 2024 Jan 23;121(4):e2313677121. doi: 10.1073/pnas.2313677121. Epub 2024 Jan 19. Proc Natl Acad Sci U S A. 2024. PMID: 38241435 Free PMC article.
References
-
- Abrahamian P., Hammond R. W., Hammond J. (2020). Plant virus-derived vectors: applications in agricultural and medical biotechnology. Annu. Rev. Virol. 7 513–535. - PubMed
-
- Akerström B., Björck L. (1986). A physicochemical study of protein G, a molecule with unique immunoglobulin G-binding properties. J. Biol. Chem. 261 10240–10247. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

