Composition and Diversity of Soil Microbial Community Associated With Land Use Types in the Agro-Pastoral Area in the Upper Yellow River Basin
- PMID: 35548288
- PMCID: PMC9082682
- DOI: 10.3389/fpls.2022.819661
Composition and Diversity of Soil Microbial Community Associated With Land Use Types in the Agro-Pastoral Area in the Upper Yellow River Basin
Abstract
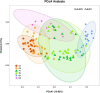
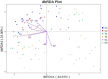
The microorganisms of soil are sensitive to their living microenvironment, and their community structure and function will change with the environmental conditions. In the agro-pastoral area on the Qinghai-Tibet Plateau, revealing the diversity of the soil microbial communities and its response to different soil physicochemical properties and environmental factors are important for ecosystem management. The microbial (bacteria and archaea) community composition and diversity under different land use types (cultivated land, grazing grassland and planted forest) were analyzed by 16S rRNA (V4 region) method in a typical agro-pastoral region in the upper Yellow River basin. Also, the soil nutrients were studied and correlated with the microbial community. The results showed that the soil nutrient contents in grassland were low, but the available nutrients were relatively high. There was a great spatial variability under different distances to the river. The microbial community diversity was lower in the grassland than the cultivated land and forest land closer to the river. For all land uses, the dominant phyla of soil microorganisms included Proteobacteria, Actinobacteria, and Bacteroidetes, while the abundance of Clostridia was significantly higher than that of the other groups, indicating that Clostridia dominated the Firmicutes and affected soil microbial community composition. The linear discriminant analysis (LDA) effect size (LefSe) analysis showed different biomarkers were more abundant in grassland than other land use types, suggesting that the structure and diversity of soil microorganisms in grassland were significantly different compared with cultivated land and forest land. The distance-based redundancy analysis (db-RDA) results showed that the total phosphorus (TP) and calcium (Ca) were the key environmental factors affecting the diversity and abundance of the soil microbial community in cultivated land and forestland, respectively. However, the microbial diversity in grassland was more related to spatial distance of the river. These results provided a theoretical basis for the changes in the composition, structure, and function of soil microbial communities in agro-pastoral areas.
Keywords: agro–pastoral area; microbial community composition; soil microbial diversity; soil property; spatial distribution.
Copyright © 2022 Liu, Sun, Shi, Liu, Wang, Dong and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Avidano L., Gamalero E., Cossa G. P., Carraro E. (2005). Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30 21–33. 10.1016/j.apsoil.2005.01.003 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

