Small Molecule Inhibitor Targeting CDT1/Geminin Protein Complex Promotes DNA Damage and Cell Death in Cancer Cells
- PMID: 35548337
- PMCID: PMC9083542
- DOI: 10.3389/fphar.2022.860682
Small Molecule Inhibitor Targeting CDT1/Geminin Protein Complex Promotes DNA Damage and Cell Death in Cancer Cells
Abstract
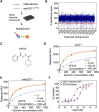
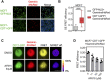
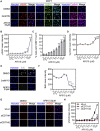
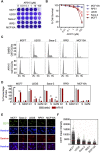
DNA replication initiation requires the loading of MCM2-7 complexes at the origins of replication during G1. Replication licensing renders chromatin competent for DNA replication and its tight regulation is essential to prevent aberrant DNA replication and genomic instability. CDT1 is a critical factor of licensing and its activity is controlled by redundant mechanisms, including Geminin, a protein inhibitor of CDT1. Aberrant CDT1 and Geminin expression have been shown to promote tumorigenesis in vivo and are also evident in multiple human tumors. In this study, we developed an in vitro AlphaScreen™ high-throughput screening (HTS) assay for the identification of small-molecule inhibitors targeting the CDT1/Geminin protein complex. Biochemical characterization of the most potent compound, AF615, provided evidence of specific, dose-dependent inhibition of Geminin binding to CDT1 both in-vitro and in cells. Moreover, compound AF615 induces DNA damage, inhibits DNA synthesis and reduces viability selectively in cancer cell lines, and this effect is CDT1-dependent. Taken together, our data suggest that AF615 may serve as a useful compound to elucidate the role of CDT1/Geminin protein complex in replication licensing and origin firing as well as a scaffold for further medicinal chemistry optimisation.
Keywords: AlphaScreen; CDT1; Geminin; cancer; high-throughput screening; small molecule inhibitor.
Copyright © 2022 Karantzelis, Petropoulos, De Marco, Egan, Fish, Christodoulou, Will, Lewis, Perrakis, Lygerou and Taraviras.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

