Identification of the cytochrome P450 enzymes involved in the oxidative metabolism of trantinterol using ultra high-performance liquid chromatography coupled with tandem mass spectrometry
- PMID: 35548639
- PMCID: PMC9086916
- DOI: 10.1039/c8ra06219f
Identification of the cytochrome P450 enzymes involved in the oxidative metabolism of trantinterol using ultra high-performance liquid chromatography coupled with tandem mass spectrometry
Abstract
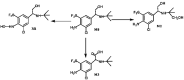
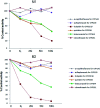
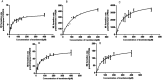
Trantinterol is a novel β2-adrenoceptor agonist used for the treatment of asthma. This study aimed to identify the cytochrome P450 enzymes responsible for the metabolism of trantinterol to form 4-hydroxylamine trantinterol (M1) and tert-butyl hydroxylated trantinterol (M2), which was achieved using the chemical inhibition study, followed by the metabolism study of trantinterol in a panel of recombinant CYPs, as well as the kinetic study with the appropriate cDNA-expressed P450 enzymes. A highly selective and sensitive ultra high-performance liquid chromatography tandem mass spectrometry method was developed and validated for the simultaneous determination of M1 and M2. The inhibition study suggested that CYP2C19 and CYP3A4/5 were involved in the formation of M1 and M2, and CYP2D6 only contributed to the formation of M1. Assays with cDNA-expressed CYP enzymes further showed that the relative contributions of P450 isoforms were 2C19 > 3A4 > 2D6 > 2E1 for the formation of M1, and 3A4 > 2C19 > 2D6 for the formation of M2. The enzyme kinetic analysis was then performed in CYP2C19, CYP2D6 and CYP3A4. The kinetic parameters were determined and normalized with respect to the human hepatic microsomal P450 isoform concentrations. All the results support the conclusion that CYP3A4 and CYP2C19 are the major enzymes responsible for formation of M1 and M2, while CYP2D6 and CYP2E1 also engaged to a lesser degree. The results imply that potential drug-drug interactions may be noticed when trantinterol is used with CYP2C19 and CYP3A4 inducers or inhibitors, and we should pay attention to this phenomenon in clinical study.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Wang Y. J. Lu X. M. Jiang K. Xiong Z. L. Cheng M. S. Li F. M. Biomed. Chromatogr. 2010;24:274. - PubMed
LinkOut - more resources
Full Text Sources

