Elucidation of the structural stability and dynamics of heterogeneous intermediate ensembles in unfolding pathway of the N-terminal domain of TDP-43
- PMID: 35548664
- PMCID: PMC9088055
- DOI: 10.1039/c8ra03368d
Elucidation of the structural stability and dynamics of heterogeneous intermediate ensembles in unfolding pathway of the N-terminal domain of TDP-43
Abstract
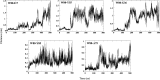
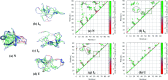
The N-terminal domain of the RNA binding protein TDP-43 (NTD) is essential to both physiology and proteinopathy; however, elucidation of its folding/unfolding still remains a major quest. In this study, we have investigated the biophysical behavior of intermediate ensembles employing all-atom molecular dynamics simulations in 8 M urea accelerated with high temperatures to achieve unfolded states in a confined computation time. The cumulative results of the 2.75 μs simulations show that unfolding of the NTD at 350 K evolves through different stable and meta-stable intermediate states. The free-energy landscape reveals two meta-stable intermediates (IN and IU) stabilized by non-native interactions, which are largely hydrophilic and highly energetically frustrated. A single buried tryptophan residue, W80, undergoes solvent exposure to different extents during unfolding; this suggests a structurally heterogeneous population of intermediate ensembles. Furthermore, the structure properties of the IN state show a resemblance to the molten globule (MG) state with most of the secondary structures intact. The unfolding of the NTD is initiated by the loss of β-strands, and the unfolded (U) states exhibit a population of non-native α-helices. These non-native unfolded intermediate ensembles may mediate protein oligomerization, leading to the formation of pathological, irreversible aggregates, characteristics of disease pathogenesis.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Neumann M. Sampathu D. M. Kwong L. K. Truax A. C. Micsenyi M. C. Chou T. T. Bruce J. Schuck T. Grossman M. Clark C. M. McCluskey L. F. Miller B. L. Masliah E. Mackenzie I. R. Feldman H. Feiden W. Kretzschmar H. A. Trojanowski J. Q. Lee V. M. Science. 2006;314:130–133. doi: 10.1126/science.1134108. - DOI - PubMed
LinkOut - more resources
Full Text Sources

