Computational Insights of Unfolding of N-Terminal Domain of TDP-43 Reveal the Conformational Heterogeneity in the Unfolding Pathway
- PMID: 35548668
- PMCID: PMC9083116
- DOI: 10.3389/fnmol.2022.822863
Computational Insights of Unfolding of N-Terminal Domain of TDP-43 Reveal the Conformational Heterogeneity in the Unfolding Pathway
Abstract
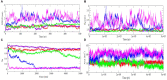
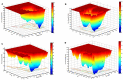
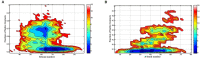
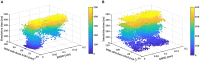
TDP-43 proteinopathies is a disease hallmark that characterizes amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). The N-terminal domain of TDP-43 (NTD) is important to both TDP-43 physiology and TDP-43 proteinopathy. However, its folding and dimerization process is still poorly characterized. In the present study, we have investigated the folding/unfolding of NTD employing all-atom molecular dynamics (MD) simulations in 8 M dimethylsulfoxide (DMSO) at high temperatures. The MD results showed that the unfolding of the NTD at high temperature evolves through the formation of a number of conformational states differing in their stability and free energy. The presence of structurally heterogeneous population of intermediate ensembles was further characterized by the different extents of solvent exposure of Trp80 during unfolding. We suggest that these non-natives unfolded intermediate ensembles may facilitate NTD oligomerization and subsequently TDP-43 oligomerization, which might lead to the formation of irreversible pathological aggregates, characteristics of disease pathogenesis.
Keywords: DMSO; NTD; TDP-43; conformational heterogeneity; unfolding intermediates.
Copyright © 2022 Li, Singh, Kashav, Yang, Sharma, Lynn, Prasad, Prakash and Kumar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Detection of an Intermediate in the Unfolding Process of the N-Terminal Domain of TDP-43.ACS Omega. 2025 Feb 5;10(6):5616-5633. doi: 10.1021/acsomega.4c08617. eCollection 2025 Feb 18. ACS Omega. 2025. PMID: 39989811 Free PMC article.
-
Elucidation of the structural stability and dynamics of heterogeneous intermediate ensembles in unfolding pathway of the N-terminal domain of TDP-43.RSC Adv. 2018 May 30;8(35):19835-19845. doi: 10.1039/c8ra03368d. eCollection 2018 May 25. RSC Adv. 2018. PMID: 35548664 Free PMC article.
-
Comparative analysis of thermal unfolding simulations of RNA recognition motifs (RRMs) of TAR DNA-binding protein 43 (TDP-43).J Biomol Struct Dyn. 2019 Jan;37(1):178-194. doi: 10.1080/07391102.2017.1422026. Epub 2018 Jan 10. J Biomol Struct Dyn. 2019. PMID: 29279008
-
[FTLD/ALS as TDP-43 proteinopathies].Rinsho Shinkeigaku. 2010 Nov;50(11):1022-4. doi: 10.5692/clinicalneurol.50.1022. Rinsho Shinkeigaku. 2010. PMID: 21921552 Review. Japanese.
-
Disease animal models of TDP-43 proteinopathy and their pre-clinical applications.Int J Mol Sci. 2013 Oct 9;14(10):20079-111. doi: 10.3390/ijms141020079. Int J Mol Sci. 2013. PMID: 24113586 Free PMC article. Review.
Cited by
-
Detection of an Intermediate in the Unfolding Process of the N-Terminal Domain of TDP-43.ACS Omega. 2025 Feb 5;10(6):5616-5633. doi: 10.1021/acsomega.4c08617. eCollection 2025 Feb 18. ACS Omega. 2025. PMID: 39989811 Free PMC article.
-
Expression analysis and mapping of Viral-Host Protein interactions of Poxviridae suggests a lead candidate molecule targeting Mpox.BMC Infect Dis. 2024 May 10;24(1):483. doi: 10.1186/s12879-024-09332-x. BMC Infect Dis. 2024. PMID: 38730352 Free PMC article.
-
Importin α/β and the tug of war to keep TDP-43 in solution: quo vadis?Bioessays. 2022 Dec;44(12):e2200181. doi: 10.1002/bies.202200181. Epub 2022 Oct 17. Bioessays. 2022. PMID: 36253101 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

