Increased SARS-CoV-2 Infection, Protease, and Inflammatory Responses in Chronic Obstructive Pulmonary Disease Primary Bronchial Epithelial Cells Defined with Single-Cell RNA Sequencing
- PMID: 35549656
- PMCID: PMC9799113
- DOI: 10.1164/rccm.202108-1901OC
Increased SARS-CoV-2 Infection, Protease, and Inflammatory Responses in Chronic Obstructive Pulmonary Disease Primary Bronchial Epithelial Cells Defined with Single-Cell RNA Sequencing
Abstract
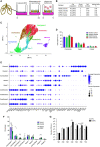
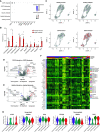
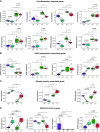
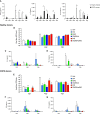
Rationale: Patients with chronic obstructive pulmonary disease (COPD) develop more severe coronavirus disease (COVID-19); however, it is unclear whether they are more susceptible to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and what mechanisms are responsible for severe disease. Objectives: To determine whether SARS-CoV-2 inoculated primary bronchial epithelial cells (pBECs) from patients with COPD support greater infection and elucidate the effects and mechanisms involved. Methods: We performed single-cell RNA sequencing analysis on differentiated pBECs from healthy subjects and patients with COPD 7 days after SARS-CoV-2 inoculation. We correlated changes with viral titers, proinflammatory responses, and IFN production. Measurements and Main Results: Single-cell RNA sequencing revealed that COPD pBECs had 24-fold greater infection than healthy cells, which was supported by plaque assays. Club/goblet and basal cells were the predominant populations infected and expressed mRNAs involved in viral replication. Proteases involved in SARS-CoV-2 entry/infection (TMPRSS2 and CTSB) were increased, and protease inhibitors (serpins) were downregulated more so in COPD. Inflammatory cytokines linked to COPD exacerbations and severe COVID-19 were increased, whereas IFN responses were blunted. Coexpression analysis revealed a prominent population of club/goblet cells with high type 1/2 IFN responses that were important drivers of immune responses to infection in both healthy and COPD pBECs. Therapeutic inhibition of proteases and inflammatory imbalances reduced viral titers and cytokine responses, particularly in COPD pBECs. Conclusions: COPD pBECs are more susceptible to SARS-CoV-2 infection because of increases in coreceptor expression and protease imbalances and have greater inflammatory responses. A prominent cluster of IFN-responsive club/goblet cells emerges during infection, which may be important drivers of immunity. Therapeutic interventions suppress SARS-CoV-2 replication and consequent inflammation.
Keywords: COPD; COVID-19; interferon; protease; single-cell RNA sequencing.
Figures





Comment in
-
The Serpin-tine Search for Factors Associated with COVID-19 Severity in Patients with Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2022 Sep 15;206(6):657-658. doi: 10.1164/rccm.202205-0956ED. Am J Respir Crit Care Med. 2022. PMID: 35612929 Free PMC article. No abstract available.
References
-
- O’Driscoll M, Ribeiro Dos Santos G, Wang L, Cummings DAT, Azman AS, Paireau J, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature . 2021;590:140–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

