A novel hypoxia-driven gene signature that can predict the prognosis of hepatocellular carcinoma
- PMID: 35549979
- PMCID: PMC9276011
- DOI: 10.1080/21655979.2022.2073943
A novel hypoxia-driven gene signature that can predict the prognosis of hepatocellular carcinoma
Abstract
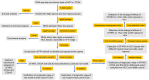
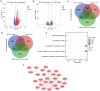
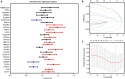
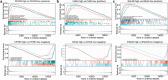
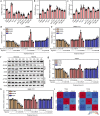
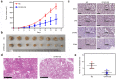
Hypoxia environment exists in already started hepatocellular carcinoma (HCC) and promotes its progression by driving changes in the gene expression profiles of cells. However, the status of hypoxia-driven genes in HCC is largely unknown. In the present study, 368 HCC tissues from The Cancer Genome Atlas were divided into high and low hypoxia groups according to their hypoxia signatures. A total of 1,142 differentially expressed genes (DEGs) were identified between the two groups, and 34 of these DEGs were highly expressed in HCC tissues compared with adjacent tissues, especially in HCC tissues from patients with stage III-IV HCC. After constructing a protein-protein interaction network and applying the least absolute shrinkage and selection operator Cox regression method for 34 DEGs, a three-gene signature (complement factor H related 3 [CFHR3], egl-9 family hypoxia inducible factor 3 [EGLN3], and chromogranin A [CHGA]) was constructed and had prognostic value to predicted outcome of patients with HCC. This three-gene signature was suitable for classifying patients with HCC in the International Cancer Genome Consortium. CFHR3 shows remarkable diagnostic value in HCC. Hypoxia decreased CFHR3 expression, but increased HCC cell proliferation and motility. Overexpression of CFHR3 in HCC cells under hypoxia reversed the stimulatory effects of hypoxia and suppressed cell proliferation and metastasis in vivo. In conclusion, we identified a novel hypoxia-driven gene signature (CFHR3, EGLN3, and CHGA) for reliable prognostic prediction of HCC, and demonstrated that overexpression of CFHR3 may be a potential strategy to overcome hypoxia and treat HCC.
Keywords: CFHR3; HCC; Hypoxia-driven gene; prognostic prediction.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
