Comparative whole-genome transcriptome analysis in renal cell populations reveals high tissue specificity of MAPK/ERK targets in embryonic kidney
- PMID: 35550069
- PMCID: PMC9102746
- DOI: 10.1186/s12915-022-01309-z
Comparative whole-genome transcriptome analysis in renal cell populations reveals high tissue specificity of MAPK/ERK targets in embryonic kidney
Abstract
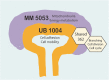
Background: MAPK/ERK signaling is a well-known mediator of extracellular stimuli controlling intracellular responses to growth factors and mechanical cues. The critical requirement of MAPK/ERK signaling for embryonic stem cell maintenance is demonstrated, but specific functions in progenitor regulation during embryonic development, and in particular kidney development remain largely unexplored. We previously demonstrated MAPK/ERK signaling as a key regulator of kidney growth through branching morphogenesis and normal nephrogenesis where it also regulates progenitor expansion. Here, we performed RNA sequencing-based whole-genome expression analysis to identify transcriptional MAPK/ERK targets in two distinct renal populations: the ureteric bud epithelium and the nephron progenitors.
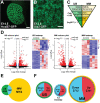
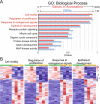
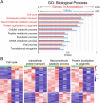
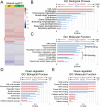
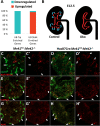
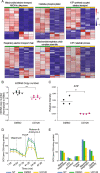
Results: Our analysis revealed a large number (5053) of differentially expressed genes (DEGs) in nephron progenitors and significantly less (1004) in ureteric bud epithelium, reflecting likely heterogenicity of cell types. The data analysis identified high tissue-specificity, as only a fraction (362) of MAPK/ERK targets are shared between the two tissues. Tissue-specific MAPK/ERK targets participate in the regulation of mitochondrial energy metabolism in nephron progenitors, which fail to maintain normal mitochondria numbers in the MAPK/ERK-deficient tissue. In the ureteric bud epithelium, a dramatic decline in progenitor-specific gene expression was detected with a simultaneous increase in differentiation-associated genes, which was not observed in nephron progenitors. Our experiments in the genetic model of MAPK/ERK deficiency provide evidence that MAPK/ERK signaling in the ureteric bud maintains epithelial cells in an undifferentiated state. Interestingly, the transcriptional targets shared between the two tissues studied are over-represented by histone genes, suggesting that MAPK/ERK signaling regulates cell cycle progression and stem cell maintenance through chromosome condensation and nucleosome assembly.
Conclusions: Using tissue-specific MAPK/ERK inactivation and RNA sequencing in combination with experimentation in embryonic kidneys, we demonstrate here that MAPK/ERK signaling maintains ureteric bud tip cells, suggesting a regulatory role in collecting duct progenitors. We additionally deliver new mechanistic information on how MAPK/ERK signaling regulates progenitor maintenance through its effects on chromatin accessibility and energy metabolism.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

