Peptide-dependent tuning of major histocompatibility complex motional properties and the consequences for cellular immunity
- PMID: 35550277
- PMCID: PMC10052791
- DOI: 10.1016/j.coi.2022.102184
Peptide-dependent tuning of major histocompatibility complex motional properties and the consequences for cellular immunity
Abstract
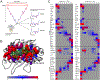
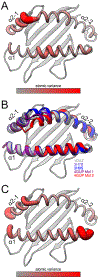
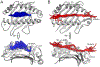
T cell receptors (TCRs) and other receptors of the immune system recognize peptides presented by class I or class II major histocompatibility complex (MHC) proteins. Although we generally distinguish between the MHC protein and its peptide, at an atomic level the two form a structural composite, which allows peptides to influence MHC properties and vice versa. One consequence is the peptide-dependent tuning of MHC structural dynamics, which contributes to protein structural adaptability and influences how receptors identify and bind targets. Peptide-dependent tuning of MHC protein dynamics can impact processes such as antigenicity, TCR cross-reactivity, and T cell repertoire selection. Motional tuning extends beyond the binding groove, influencing peptide selection and exchange, as well as interactions with other immune receptors. Here, we review recent findings showing how peptides can affect the dynamic and adaptable nature of MHC proteins. We highlight consequences for immunity and demonstrate how MHC proteins have evolved to be highly sensitive dynamic reporters, with broad immunological consequences.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interest
The authors declare no conflicts of interest.
Figures
References
-
- Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J: T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annual Review of Immunology 2015, 33:169–200. - PubMed
-
- Miles JJ, McCluskey J, Rossjohn J, Gras S: Understanding the complexity and malleability of T-cell recognition. Immunol Cell Biol 2015, 93:433–441. - PubMed
-
- Kass I, Buckle AM, Borg NA: Understanding the structural dynamics of TCR-pMHC interactions. Trends in Immunology 2014, 35:604–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

