CDK12 promotes tumorigenesis but induces vulnerability to therapies inhibiting folate one-carbon metabolism in breast cancer
- PMID: 35550508
- PMCID: PMC9098894
- DOI: 10.1038/s41467-022-30375-8
CDK12 promotes tumorigenesis but induces vulnerability to therapies inhibiting folate one-carbon metabolism in breast cancer
Abstract
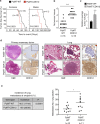
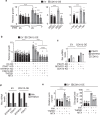
Cyclin-dependent kinase 12 (CDK12) overexpression is implicated in breast cancer, but whether it has a primary or only a cooperative tumorigenic role is unclear. Here, we show that transgenic CDK12 overexpression in the mouse mammary gland per se is sufficient to drive the emergence of multiple and multifocal tumors, while, in cooperation with known oncogenes, it promotes earlier tumor onset and metastasis. Integrative transcriptomic, metabolomic and functional data reveal that hyperactivation of the serine-glycine-one-carbon network is a metabolic hallmark inherent to CDK12-induced tumorigenesis. Consistently, in retrospective patient cohort studies and in patient-derived xenografts, CDK12-overexpressing breast tumors show positive response to methotrexate-based chemotherapy targeting CDK12-induced metabolic alterations, while being intrinsically refractory to other types of chemotherapy. In a retrospective analysis of hormone receptor-negative and lymph node-positive breast cancer patients randomized in an adjuvant phase III trial to 1-year low-dose metronomic methotrexate-based chemotherapy or no maintenance chemotherapy, a high CDK12 status predicts a dramatic reduction in distant metastasis rate in the chemotherapy-treated vs. not-treated arm. Thus, by coupling tumor progression with metabolic reprogramming, CDK12 creates an actionable vulnerability for breast cancer therapy and might represent a suitable companion biomarker for targeted antimetabolite therapies in human breast cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

