The early-life exposome modulates the effect of polymorphic inversions on DNA methylation
- PMID: 35550596
- PMCID: PMC9098634
- DOI: 10.1038/s42003-022-03380-2
The early-life exposome modulates the effect of polymorphic inversions on DNA methylation
Abstract
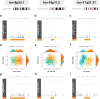
Polymorphic genomic inversions are chromosomal variants with intrinsic variability that play important roles in evolution, environmental adaptation, and complex traits. We investigated the DNA methylation patterns of three common human inversions, at 8p23.1, 16p11.2, and 17q21.31 in 1,009 blood samples from children from the Human Early Life Exposome (HELIX) project and in 39 prenatal heart tissue samples. We found inversion-state specific methylation patterns within and nearby flanking each inversion region in both datasets. Additionally, numerous inversion-exposure interactions on methylation levels were identified from early-life exposome data comprising 64 exposures. For instance, children homozygous at inv-8p23.1 and higher meat intake were more susceptible to TDH hypermethylation (P = 3.8 × 10-22); being the inversion, exposure, and gene known risk factors for adult obesity. Inv-8p23.1 associated hypermethylation of GATA4 was also detected across numerous exposures. Our data suggests that the pleiotropic influence of inversions during development and lifetime could be substantially mediated by allele-specific methylation patterns which can be modulated by the exposome.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

