UBR5 targets tumor suppressor CDC73 proteolytically to promote aggressive breast cancer
- PMID: 35551175
- PMCID: PMC9098409
- DOI: 10.1038/s41419-022-04914-6
UBR5 targets tumor suppressor CDC73 proteolytically to promote aggressive breast cancer
Abstract
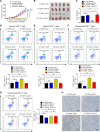
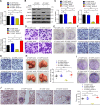
UBR5, a HECT-domain E3 ubiquitin ligase, is an attractive therapeutic target for aggressive breast cancers. Defining the substrates of UBR5 is crucial for scientific understanding and clinical intervention. Here, we demonstrate that CDC73, a component of the RNA polymerase II-associated factor 1 complex, is a key substrate that impedes UBR5's profound tumorigenic and metastatic activities in triple-negative breast cancer (TNBC) via mechanisms of regulating the expression of β-catenin and E-cadherin, tumor cell apoptosis and CD8+ T cell infiltration. Expression of CDC73 is also negatively associated with the progression of breast cancer patients. Moreover, we show that UBR5 destabilizes CDC73 by polyubiquitination at Lys243, Lys247, and Lys257 in a non-canonical manner that is dependent on the non-phosphorylation state of CDC73 at Ser465. CDC73 could serve as a molecular switch to modulate UBR5's pro-tumor activities and may provide a potential approach to developing breast cancer therapeutic interventions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Targeting UBR5 inhibits postsurgical breast cancer lung metastases by inducing CDC73 and p53 mediated apoptosis.Int J Cancer. 2024 Feb 15;154(4):723-737. doi: 10.1002/ijc.34769. Epub 2023 Oct 19. Int J Cancer. 2024. PMID: 37855385 Free PMC article.
-
E3 Ubiquitin Ligase UBR5 Drives the Growth and Metastasis of Triple-Negative Breast Cancer.Cancer Res. 2017 Apr 15;77(8):2090-2101. doi: 10.1158/0008-5472.CAN-16-2409. Epub 2017 Mar 22. Cancer Res. 2017. PMID: 28330927
-
UBR5 promotes tumor immune evasion through enhancing IFN-γ-induced PDL1 transcription in triple negative breast cancer.Theranostics. 2022 Jul 4;12(11):5086-5102. doi: 10.7150/thno.74989. eCollection 2022. Theranostics. 2022. PMID: 35836797 Free PMC article.
-
UBR5 in Tumor Biology: Exploring Mechanisms of Immune Regulation and Possible Therapeutic Implications in MPNST.Cancers (Basel). 2025 Jan 7;17(2):161. doi: 10.3390/cancers17020161. Cancers (Basel). 2025. PMID: 39857943 Free PMC article. Review.
-
Functional significance and therapeutic implication of ring-type E3 ligases in colorectal cancer.Oncogene. 2018 Jan 11;37(2):148-159. doi: 10.1038/onc.2017.313. Epub 2017 Sep 18. Oncogene. 2018. PMID: 28925398 Free PMC article. Review.
Cited by
-
SUB1 promotes colorectal cancer metastasis by activating NF-κB signaling via UBR5-mediated ubiquitination of UBXN1.Sci China Life Sci. 2024 Jun;67(6):1199-1211. doi: 10.1007/s11427-023-2429-5. Epub 2024 Jan 10. Sci China Life Sci. 2024. PMID: 38240906
-
lncRNA Ubr5 promotes BMSCs apoptosis and inhibits their proliferation and osteogenic differentiation in weightless bone loss.Front Cell Dev Biol. 2025 Apr 2;13:1543929. doi: 10.3389/fcell.2025.1543929. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40241795 Free PMC article.
-
Structural snapshots along K48-linked ubiquitin chain formation by the HECT E3 UBR5.Nat Chem Biol. 2024 Feb;20(2):190-200. doi: 10.1038/s41589-023-01414-2. Epub 2023 Aug 24. Nat Chem Biol. 2024. PMID: 37620400 Free PMC article.
-
A review: targeting UBR5 domains to mediate emerging roles and mechanisms - chance or necessity?Int J Surg. 2024 Aug 1;110(8):4947-4964. doi: 10.1097/JS9.0000000000001541. Int J Surg. 2024. PMID: 38701508 Free PMC article. Review.
-
Whole‑exome sequencing insights into synchronous bilateral breast cancer with discordant molecular subtypes.Oncol Lett. 2024 Oct 8;28(6):595. doi: 10.3892/ol.2024.14728. eCollection 2024 Dec. Oncol Lett. 2024. PMID: 39430730 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

