Large differences in carbohydrate degradation and transport potential among lichen fungal symbionts
- PMID: 35551185
- PMCID: PMC9098629
- DOI: 10.1038/s41467-022-30218-6
Large differences in carbohydrate degradation and transport potential among lichen fungal symbionts
Abstract
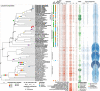
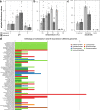
Lichen symbioses are thought to be stabilized by the transfer of fixed carbon from a photosynthesizing symbiont to a fungus. In other fungal symbioses, carbohydrate subsidies correlate with reductions in plant cell wall-degrading enzymes, but whether this is true of lichen fungal symbionts (LFSs) is unknown. Here, we predict genes encoding carbohydrate-active enzymes (CAZymes) and sugar transporters in 46 genomes from the Lecanoromycetes, the largest extant clade of LFSs. All LFSs possess a robust CAZyme arsenal including enzymes acting on cellulose and hemicellulose, confirmed by experimental assays. However, the number of genes and predicted functions of CAZymes vary widely, with some fungal symbionts possessing arsenals on par with well-known saprotrophic fungi. These results suggest that stable fungal association with a phototroph does not in itself result in fungal CAZyme loss, and lends support to long-standing hypotheses that some lichens may augment fixed CO2 with carbon from external sources.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Schwendener S. Die Algentypen der Flechtengonidien. Basel: Universitaetsbuchdruckerei; 1869.
-
- Drew EA, Smith DC. Studies in the physiology of lichens. VII. The physiology of the Nostoc symbiont of Peltigera polydactyla compared with cultured and free-living forms. N. Phytologist. 1967;66:379–388. doi: 10.1111/j.1469-8137.1967.tb06017.x. - DOI
-
- Richardson DL, Hill DJ, Smith D. Lichen Physiology - XI. The role of the alga in determining the pattern of carbohydrate movement between lichen symbionts. N. Phytologist. 1968;67:469–486. doi: 10.1111/j.1469-8137.1968.tb05476.x. - DOI
-
- Smith, D. C. Mechanisms of nutrient movement between the lichen symbionts. In Cellular interactions in symbiosis and parasitism (eds Cook, C. B., Pappas, P. W. & Rudolph, E. D.). 197–227 (Ohio State University Press, 1980).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

