Structural basis of ion - substrate coupling in the Na+-dependent dicarboxylate transporter VcINDY
- PMID: 35551191
- PMCID: PMC9098524
- DOI: 10.1038/s41467-022-30406-4
Structural basis of ion - substrate coupling in the Na+-dependent dicarboxylate transporter VcINDY
Abstract
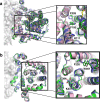
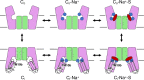
The Na+-dependent dicarboxylate transporter from Vibrio cholerae (VcINDY) is a prototype for the divalent anion sodium symporter (DASS) family. While the utilization of an electrochemical Na+ gradient to power substrate transport is well established for VcINDY, the structural basis of this coupling between sodium and substrate binding is not currently understood. Here, using a combination of cryo-EM structure determination, succinate binding and site-directed cysteine alkylation assays, we demonstrate that the VcINDY protein couples sodium- and substrate-binding via a previously unseen cooperative mechanism by conformational selection. In the absence of sodium, substrate binding is abolished, with the succinate binding regions exhibiting increased flexibility, including HPinb, TM10b and the substrate clamshell motifs. Upon sodium binding, these regions become structurally ordered and create a proper binding site for the substrate. Taken together, these results provide strong evidence that VcINDY's conformational selection mechanism is a result of the sodium-dependent formation of the substrate binding site.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

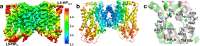


Similar articles
-
Solvent accessibility changes in a Na+-dependent C4-dicarboxylate transporter suggest differential substrate effects in a multistep mechanism.J Biol Chem. 2020 Dec 25;295(52):18524-18538. doi: 10.1074/jbc.RA120.013894. Epub 2020 Oct 21. J Biol Chem. 2020. PMID: 33087444 Free PMC article.
-
Functional characterization of a Na+-dependent dicarboxylate transporter from Vibrio cholerae.J Gen Physiol. 2014 Jun;143(6):745-59. doi: 10.1085/jgp.201311141. Epub 2014 May 12. J Gen Physiol. 2014. PMID: 24821967 Free PMC article.
-
Structure and function of the divalent anion/Na+ symporter from Vibrio cholerae and a humanized variant.Nat Commun. 2017 Apr 24;8:15009. doi: 10.1038/ncomms15009. Nat Commun. 2017. PMID: 28436435 Free PMC article.
-
[Progress in the investigation of Na+/dicarboxylate co-transporter].Sheng Li Ke Xue Jin Zhan. 2003 Jul;34(3):245-7. Sheng Li Ke Xue Jin Zhan. 2003. PMID: 14628473 Review. Chinese. No abstract available.
-
C4-dicarboxylate carriers and sensors in bacteria.Biochim Biophys Acta. 2002 Jan 17;1553(1-2):39-56. doi: 10.1016/s0005-2728(01)00233-x. Biochim Biophys Acta. 2002. PMID: 11803016 Review.
Cited by
-
Large-scale experimental assessment of variant effects on the structure and function of the citrate transporter SLC13A5.Sci Adv. 2025 Jun 27;11(26):eadx3011. doi: 10.1126/sciadv.adx3011. Epub 2025 Jun 27. Sci Adv. 2025. PMID: 40577459 Free PMC article.
-
Substrate translocation and inhibition in human dicarboxylate transporter NaDC3.Nat Struct Mol Biol. 2025 Mar;32(3):502-512. doi: 10.1038/s41594-024-01433-0. Epub 2024 Dec 2. Nat Struct Mol Biol. 2025. PMID: 39622972
-
Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.Nat Commun. 2024 Jun 29;15(1):5503. doi: 10.1038/s41467-024-48921-x. Nat Commun. 2024. PMID: 38951531 Free PMC article.
-
Structural basis for the reaction cycle and transport mechanism of human Na+-sulfate cotransporter NaS1 (SLC13A1).Sci Adv. 2024 Nov 22;10(47):eado6778. doi: 10.1126/sciadv.ado6778. Epub 2024 Nov 22. Sci Adv. 2024. PMID: 39576865 Free PMC article.
-
Conformational coupling of the sialic acid TRAP transporter HiSiaQM with its substrate binding protein HiSiaP.Nat Commun. 2024 Jan 8;15(1):217. doi: 10.1038/s41467-023-44327-3. Nat Commun. 2024. PMID: 38191530 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

