Germline mutations in mitochondrial complex I reveal genetic and targetable vulnerability in IDH1-mutant acute myeloid leukaemia
- PMID: 35551192
- PMCID: PMC9098909
- DOI: 10.1038/s41467-022-30223-9
Germline mutations in mitochondrial complex I reveal genetic and targetable vulnerability in IDH1-mutant acute myeloid leukaemia
Erratum in
-
Author Correction: Germline mutations in mitochondrial complex I reveal genetic and targetable vulnerability in IDH1-mutant acute myeloid leukaemia.Nat Commun. 2022 Jul 15;13(1):4131. doi: 10.1038/s41467-022-31952-7. Nat Commun. 2022. PMID: 35840577 Free PMC article. No abstract available.
Abstract
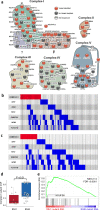
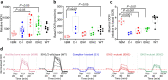
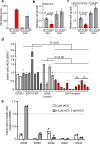
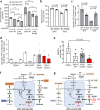
The interaction of germline variation and somatic cancer driver mutations is under-investigated. Here we describe the genomic mitochondrial landscape in adult acute myeloid leukaemia (AML) and show that rare variants affecting the nuclear- and mitochondrially-encoded complex I genes show near-mutual exclusivity with somatic driver mutations affecting isocitrate dehydrogenase 1 (IDH1), but not IDH2 suggesting a unique epistatic relationship. Whereas AML cells with rare complex I variants or mutations in IDH1 or IDH2 all display attenuated mitochondrial respiration, heightened sensitivity to complex I inhibitors including the clinical-grade inhibitor, IACS-010759, is observed only for IDH1-mutant AML. Furthermore, IDH1 mutant blasts that are resistant to the IDH1-mutant inhibitor, ivosidenib, retain sensitivity to complex I inhibition. We propose that the IDH1 mutation limits the flexibility for citrate utilization in the presence of impaired complex I activity to a degree that is not apparent in IDH2 mutant cells, exposing a mutation-specific metabolic vulnerability. This reduced metabolic plasticity explains the epistatic relationship between the germline complex I variants and oncogenic IDH1 mutation underscoring the utility of genomic data in revealing metabolic vulnerabilities with implications for therapy.
© 2022. The Author(s).
Conflict of interest statement
R.M. is on the Board of Directors of CircBio Inc., and Advisory Boards of Kodikaz Therapeutic Solutions Inc. and Syros Pharmaceuticals. R.M. is an equity holder and founder of CircBio Inc. and Pheast Therapeutics Inc.. R.M. is an inventor on several patents related to CD47 cancer immunotherapy licensed to Gilead Sciences, Inc. that are not directly related to the research in this study. The remaining authors declare no competing interests.
Figures
References
-
- Stuani L, Sarry JE. Microenvironmental aspartate preserves leukemic cells from therapy-induced metabolic collapse. Cell Metab. 2020;32:321–323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

