Structural dynamics of SARS-CoV-2 nucleocapsid protein induced by RNA binding
- PMID: 35551296
- PMCID: PMC9129039
- DOI: 10.1371/journal.pcbi.1010121
Structural dynamics of SARS-CoV-2 nucleocapsid protein induced by RNA binding
Abstract
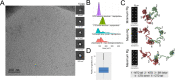
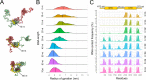
The nucleocapsid (N) protein of the SARS-CoV-2 virus, the causal agent of COVID-19, is a multifunction phosphoprotein that plays critical roles in the virus life cycle, including transcription and packaging of the viral RNA. To play such diverse roles, the N protein has two globular RNA-binding modules, the N- (NTD) and C-terminal (CTD) domains, which are connected by an intrinsically disordered region. Despite the wealth of structural data available for the isolated NTD and CTD, how these domains are arranged in the full-length protein and how the oligomerization of N influences its RNA-binding activity remains largely unclear. Herein, using experimental data from electron microscopy and biochemical/biophysical techniques combined with molecular modeling and molecular dynamics simulations, we show that, in the absence of RNA, the N protein formed structurally dynamic dimers, with the NTD and CTD arranged in extended conformations. However, in the presence of RNA, the N protein assumed a more compact conformation where the NTD and CTD are packed together. We also provided an octameric model for the full-length N bound to RNA that is consistent with electron microscopy images of the N protein in the presence of RNA. Together, our results shed new light on the dynamics and higher-order oligomeric structure of this versatile protein.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Assembly of SARS-CoV-2 nucleocapsid protein with nucleic acid.Nucleic Acids Res. 2024 Jun 24;52(11):6647-6661. doi: 10.1093/nar/gkae256. Nucleic Acids Res. 2024. PMID: 38587193 Free PMC article.
-
Modulation of biophysical properties of nucleocapsid protein in the mutant spectrum of SARS-CoV-2.Elife. 2024 Jun 28;13:RP94836. doi: 10.7554/eLife.94836. Elife. 2024. PMID: 38941236 Free PMC article.
-
A core network in the SARS-CoV-2 nucleocapsid NTD mediates structural integrity and selective RNA-binding.Nat Commun. 2024 Dec 9;15(1):10656. doi: 10.1038/s41467-024-55024-0. Nat Commun. 2024. PMID: 39653699 Free PMC article.
-
Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization.J Biol Chem. 2024 Nov;300(11):107834. doi: 10.1016/j.jbc.2024.107834. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39343000 Free PMC article. Review.
-
Unraveling the role of the nucleocapsid protein in SARS-CoV-2 pathogenesis: From viral life cycle to vaccine development.Int J Biol Macromol. 2024 Nov;279(Pt 2):135201. doi: 10.1016/j.ijbiomac.2024.135201. Epub 2024 Aug 30. Int J Biol Macromol. 2024. PMID: 39216563 Review.
Cited by
-
Liquid-liquid phase separation of nucleocapsid proteins during SARS-CoV-2 and HIV-1 replication.Cell Rep. 2023 Jan 31;42(1):111968. doi: 10.1016/j.celrep.2022.111968. Epub 2022 Dec 26. Cell Rep. 2023. PMID: 36640305 Free PMC article. Review.
-
The Role of TDP-43 in SARS-CoV-2-Related Neurodegenerative Changes.Viruses. 2025 May 19;17(5):724. doi: 10.3390/v17050724. Viruses. 2025. PMID: 40431734 Free PMC article. Review.
-
An RNA-Scaffold Protein Subunit Vaccine for Nasal Immunization.Vaccines (Basel). 2023 Sep 29;11(10):1550. doi: 10.3390/vaccines11101550. Vaccines (Basel). 2023. PMID: 37896953 Free PMC article.
-
Characterization of the binding features between SARS-CoV-2 5'-proximal transcripts of genomic RNA and nucleocapsid proteins.RNA Biol. 2025 Dec;22(1):1-16. doi: 10.1080/15476286.2025.2471643. Epub 2025 Mar 12. RNA Biol. 2025. PMID: 40077853 Free PMC article. Review.
-
Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.Sci Rep. 2022 Nov 2;12(1):18500. doi: 10.1038/s41598-022-22576-4. Sci Rep. 2022. PMID: 36323732 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

