Lipid pathway dysfunction is prevalent in patients with Parkinson's disease
- PMID: 35551349
- PMCID: PMC9586542
- DOI: 10.1093/brain/awac176
Lipid pathway dysfunction is prevalent in patients with Parkinson's disease
Abstract
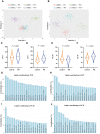
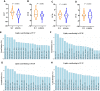
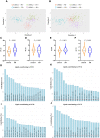
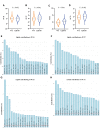
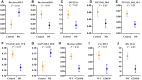
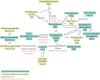
Many genetic risk factors for Parkinson's disease have lipid-related functions and lipid-modulating drugs such as statins may be protective against Parkinson's disease. Moreover, the hallmark Parkinson's disease pathological protein, α-synuclein, has lipid membrane function and pathways dysregulated in Parkinson's disease such as the endosome-lysosome system and synaptic signalling rely heavily on lipid dynamics. Despite the potential role for lipids in Parkinson's disease, most research to date has been protein-centric, with large-scale, untargeted serum and CSF lipidomic comparisons between genetic and idiopathic Parkinson's disease and neurotypical controls limited. In particular, the extent to which lipid dysregulation occurs in mutation carriers of one of the most common Parkinson's disease risk genes, LRRK2, is unclear. Further, the functional lipid pathways potentially dysregulated in idiopathic and LRRK2 mutation Parkinson's disease are underexplored. To better determine the extent of lipid dysregulation in Parkinson's disease, untargeted high-performance liquid chromatography-tandem mass spectrometry was performed on serum (n = 221) and CSF (n = 88) obtained from a multi-ethnic population from the Michael J. Fox Foundation LRRK2 Clinical Cohort Consortium. The cohort consisted of controls, asymptomatic LRRK2 G2019S carriers, LRRK2 G2019S carriers with Parkinson's disease and Parkinson's disease patients without a LRRK2 mutation. Age and sex were adjusted for in analyses where appropriate. Approximately 1000 serum lipid species per participant were analysed. The main serum lipids that distinguished both Parkinson's disease patients and LRRK2 mutation carriers from controls included species of ceramide, triacylglycerol, sphingomyelin, acylcarnitine, phosphatidylcholine and lysophosphatidylethanolamine. Significant alterations in sphingolipids and glycerolipids were also reflected in Parkinson's disease and LRRK2 mutation carrier CSF, although no correlations were observed between lipids identified in both serum and CSF. Pathway analysis of altered lipid species indicated that sphingolipid metabolism, insulin signalling and mitochondrial function were the major metabolic pathways dysregulated in Parkinson's disease. Importantly, these pathways were also found to be dysregulated in serum samples from a second Parkinson's disease cohort (n = 315). Results from this study demonstrate that dysregulated lipids in Parkinson's disease generally, and in LRRK2 mutation carriers, are from functionally and metabolically related pathways. These findings provide new insight into the extent of lipid dysfunction in Parkinson's disease and therapeutics manipulating these pathways may be beneficial for Parkinson's disease patients. Moreover, serum lipid profiles may be novel biomarkers for both genetic and idiopathic Parkinson's disease.
Keywords: LRRK2; Parkinson’s disease; biomarker; lipid; sphingolipid.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures
References
-
- Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351(19):1972–1977. - PubMed
-
- de Carvalho Guimarães B, Valente Pereira AC, da Costa Rodrigues F, et al. . Glucocerebrosidase N370S and L444P mutations as risk factors for Parkinson’s disease in Brazilian patients. Parkinsonism Relat Disord. 2012;18(5):688–689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

