Phylogenetic and amino acid signature analysis of the SARS-CoV-2s lineages circulating in Tunisia
- PMID: 35552003
- PMCID: PMC9085353
- DOI: 10.1016/j.meegid.2022.105300
Phylogenetic and amino acid signature analysis of the SARS-CoV-2s lineages circulating in Tunisia
Abstract
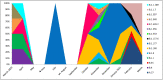
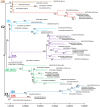
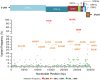
Since the beginning of the Coronavirus disease-2019 pandemic, there has been a growing interest in exploring SARS-CoV-2 genetic variation to understand the origin and spread of the pandemic, improve diagnostic methods and develop the appropriate vaccines. The objective of this study was to identify the SARS-CoV-2s lineages circulating in Tunisia and to explore their amino acid signature in order to follow their genome dynamics. Whole genome sequencing and genetic analyses of fifty-eight SARS-CoV-2 samples collected during one-year between March 2020 and March 2021 from the National Influenza Center were performed using three sampling strategies.. Multiple lineage introductions were noted during the initial phase of the pandemic, including B.4, B.1.1, B.1.428.2, B.1.540 and B.1.1.189. Subsequently, lineages B1.160 (24.2%) and B1.177 (22.4%) were dominant throughout the year. The Alpha variant (B.1.1.7 lineage) was identified in February 2021 and firstly observed in the center of our country. In addition, A clear diversity of lineages was observed in the North of the country. A total of 335 mutations including 10 deletions were found. The SARS-CoV-2 proteins ORF1ab, Spike, ORF3a, and Nucleocapsid were observed as mutation hotspots with a mutation frequency exceeding 20%. The 2 most frequent mutations, D614G in S protein and P314L in Nsp12 appeared simultaneously and are often associated with increased viral infectivity. Interestingly, deletions in coding regions causing consequent deletions of amino acids and frame shifts were identified in NSP3, NSP6, S, E, ORF7a, ORF8 and N proteins. These findings contribute to define the COVID-19 outbreak in Tunisia. Despite the country's limited resources, surveillance of SARS-CoV-2 genomic variation should be continued to control the occurrence of new variants.
Keywords: Amino acid change analysis; Amino acid signature; Coronavirus disease-2019; Lineages phylogenetic; SARS-CoV-2; Tunisia; Whole genome sequencing.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that there is no competing interest.
Figures
Similar articles
-
The extent of molecular variation in novel SARS-CoV-2 after the six-month global spread.Infect Genet Evol. 2021 Jul;91:104800. doi: 10.1016/j.meegid.2021.104800. Epub 2021 Mar 5. Infect Genet Evol. 2021. PMID: 33677109 Free PMC article.
-
Global variation in SARS-CoV-2 proteome and its implication in pre-lockdown emergence and dissemination of 5 dominant SARS-CoV-2 clades.Infect Genet Evol. 2021 Sep;93:104973. doi: 10.1016/j.meegid.2021.104973. Epub 2021 Jun 18. Infect Genet Evol. 2021. PMID: 34147651 Free PMC article.
-
Genome sequence diversity of SARS-CoV-2 obtained from clinical samples in Uzbekistan.PLoS One. 2022 Jun 27;17(6):e0270314. doi: 10.1371/journal.pone.0270314. eCollection 2022. PLoS One. 2022. PMID: 35759503 Free PMC article.
-
Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review.Eur J Med Res. 2021 Jun 8;26(1):51. doi: 10.1186/s40001-021-00524-8. Eur J Med Res. 2021. PMID: 34103090 Free PMC article.
-
A comprehensive review of current insights into the virulence factors of SARS-CoV-2.J Virol. 2025 Feb 25;99(2):e0204924. doi: 10.1128/jvi.02049-24. Epub 2025 Jan 29. J Virol. 2025. PMID: 39878471 Free PMC article. Review.
Cited by
-
Molecular characterization, phylogenetic and variation analyses of SARS-CoV-2 strains in India.Virusdisease. 2024 Sep;35(3):462-477. doi: 10.1007/s13337-024-00878-7. Epub 2024 Aug 9. Virusdisease. 2024. PMID: 39464729
-
Wastewater sequencing from a rural community enables identification of widespread adaptive mutations in a SARS-CoV-2 alpha variant.Sci Rep. 2025 May 28;15(1):18657. doi: 10.1038/s41598-025-03771-5. Sci Rep. 2025. PMID: 40437225 Free PMC article.
-
Virological Aspects of COVID-19 in Patients with Hematological Malignancies: Duration of Viral Shedding and Genetic Analysis.Viruses. 2024 Dec 31;17(1):46. doi: 10.3390/v17010046. Viruses. 2024. PMID: 39861838 Free PMC article.
-
Computational analysis of the sequence-structure relation in SARS-CoV-2 spike protein using protein contact networks.Sci Rep. 2023 Feb 17;13(1):2837. doi: 10.1038/s41598-023-30052-w. Sci Rep. 2023. PMID: 36808182 Free PMC article.
References
-
- Abid S., Ferjani S., El Moussi A., Ferjani A., Nasr M., Landolsi I., Saidi K., Gharbi H., Letaief H., Hechaichi A., Safer M., Ben Alaya N., Boubaker I.B. Assessment of sample pooling for SARS-CoV-2 molecular testing for screening of asymptomatic persons in Tunisia. Diagn. Microbiol. Infect. Dis. 2020;98(3) doi: 10.1016/j.diagmicrobio.2020.115125. - DOI - PMC - PubMed
-
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science (New York, N.Y.) 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. - DOI - PMC - PubMed
-
- Chakroun H., Ben Lasfar N., Fall S., Maha A., El Moussi A., Abid S., Rouis S., Bellazreg F., Abassi-Bakir D., Bouafif Ben Alaya N., Boutiba Ben Boubaker I., Hachfi W., Letaief A. First case of imported and confirmed COVID-19 in Tunisia. La Tunisiemedicale. 2020;98(4):258–260. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

