rRNA operon multiplicity as a bacterial genome stability insurance policy
- PMID: 35552441
- PMCID: PMC9825170
- DOI: 10.1093/nar/gkac332
rRNA operon multiplicity as a bacterial genome stability insurance policy
Abstract
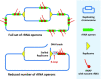
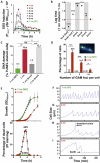
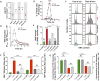
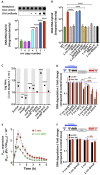
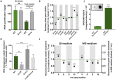
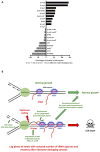
Quick growth restart after upon encountering favourable environmental conditions is a major fitness contributor in natural environment. It is widely assumed that the time required to restart growth after nutritional upshift is determined by how long it takes for cells to synthesize enough ribosomes to produce the proteins required to reinitiate growth. Here we show that a reduction in the capacity to synthesize ribosomes by reducing number of ribosomal RNA (rRNA) operons (rrn) causes a longer transition from stationary phase to growth of Escherichia coli primarily due to high mortality rates. Cell death results from DNA replication blockage and massive DNA breakage at the sites of the remaining rrn operons that become overloaded with RNA polymerases (RNAPs). Mortality rates and growth restart duration can be reduced by preventing R-loop formation and improving DNA repair capacity. The same molecular mechanisms determine the duration of the recovery phase after ribosome-damaging stresses, such as antibiotics, exposure to bile salts or high temperature. Our study therefore suggests that a major function of rrn operon multiplicity is to ensure that individual rrn operons are not saturated by RNAPs, which can result in catastrophic chromosome replication failure and cell death during adaptation to environmental fluctuations.
Plain language summary
The ability to modulate translation capacity, which resides greatly on a number of ribosomes, provides robustness in fluctuating environments. Because translation is energetically the most expensive process in cells, cells must constantly adapt the rate of ribosome production to resource availability. This is primarily achieved by regulating ribosomal RNA (rRNA) synthesis, to which ribosomal proteins synthesis is adjusted. The multiplicity of rRNA encoding operons per bacterial genome exceeds requirements for the maximal growth rates in non-stress conditions. In this study, the authors provide evidence that a major function of rRNA operon multiplicity is to ensure that individual operons are not saturated by RNA polymerases during adaptation to environmental fluctuations, which can result in catastrophic chromosome replication failure and cell death.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

