Pleiotropic Enhancers are Ubiquitous Regulatory Elements in the Human Genome
- PMID: 35552697
- PMCID: PMC9156028
- DOI: 10.1093/gbe/evac071
Pleiotropic Enhancers are Ubiquitous Regulatory Elements in the Human Genome
Abstract
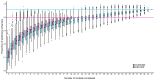
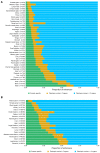
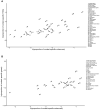
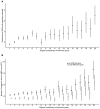
Enhancers are regulatory elements of genomes that determine spatio-temporal patterns of gene expression. The human genome contains a vast number of enhancers, which largely outnumber protein-coding genes. Historically, enhancers have been regarded as highly tissue-specific. However, recent evidence has demonstrated that many enhancers are pleiotropic, with activity in multiple developmental contexts. Yet, the extent and impact of pleiotropy remain largely unexplored. In this study we analyzed active enhancers across human organs based on the analysis of both eRNA transcription (FANTOM5 consortium data sets) and chromatin architecture (ENCODE consortium data sets). We show that pleiotropic enhancers are pervasive in the human genome and that most enhancers active in a particular organ are also active in other organs. In addition, our analysis suggests that the proportion of context-specific enhancers of a given organ is explained, at least in part, by the proportion of context-specific genes in that same organ. The notion that such a high proportion of human enhancers can be pleiotropic suggests that small regions of regulatory DNA contain abundant regulatory information and that these regions evolve under important evolutionary constraints.
Keywords: CRE; enhancer; gene regulation; human genome; non-coding DNA; pleiotropy.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Djureinovic D, et al. 2014. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 20:476–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

