Population-specific, recent positive selection signatures in cultivated Cucumis sativus L. (cucumber)
- PMID: 35554526
- PMCID: PMC9258548
- DOI: 10.1093/g3journal/jkac119
Population-specific, recent positive selection signatures in cultivated Cucumis sativus L. (cucumber)
Abstract
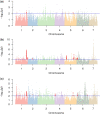
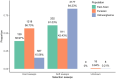
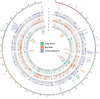
Population-specific, positive selection promotes the diversity of populations and drives local adaptations in the population. However, little is known about population-specific, recent positive selection in the populations of cultivated cucumber (Cucumis sativus L.). Based on a genomic variation map of individuals worldwide, we implemented a Fisher's combination method by combining 4 haplotype-based approaches: integrated haplotype score (iHS), number of segregating sites by length (nSL), cross-population extended haplotype homozygosity (XP-EHH), and Rsb. Overall, we detected 331, 2,147, and 3,772 population-specific, recent positive selective sites in the East Asian, Eurasian, and Xishuangbanna populations, respectively. Moreover, we found that these sites were related to processes for reproduction, response to abiotic and biotic stress, and regulation of developmental processes, indicating adaptations to their microenvironments. Meanwhile, the selective genes associated with traits of fruits were also observed, such as the gene related to the shorter fruit length in the Eurasian population and the gene controlling flesh thickness in the Xishuangbanna population. In addition, we noticed that soft sweeps were common in the East Asian and Xishuangbanna populations. Genes involved in hard or soft sweeps were related to developmental regulation and abiotic and biotic stress resistance. Our study offers a comprehensive candidate dataset of population-specific, selective signatures in cultivated cucumber populations. Our methods provide guidance for the analysis of population-specific, positive selection. These findings will help explore the biological mechanisms of adaptation and domestication of cucumber.
Keywords: Cucumis sativus L; Fisher’s combination; cultivated cucumber; population-specific; recent positive selection.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures

References
-
- Acer V, Ali S, Nathan CE, Murilo MP, Jason O, David BL.. A molecular view of plant local adaptation: incorporating stress-response networks. Annu Rev Plant Biol. 2019;70:559–583. - PubMed
-
- Agarwal PK, Gupta K, Lopato S, Agarwal P.. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot. 2017;68(9):2135–2148. - PubMed
-
- Alexandra V, Eric B, Oscar G.. Detection of selective sweeps in structured populations: a comparison of recent methods. Mol Ecol. 2015;25(1):89–103. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
