Proteomic characterization of four subtypes of M2 macrophages derived from human THP-1 cells
- PMID: 35557041
- PMCID: PMC9110321
- DOI: 10.1631/jzus.B2100930
Proteomic characterization of four subtypes of M2 macrophages derived from human THP-1 cells
Abstract
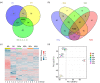
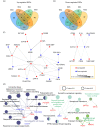
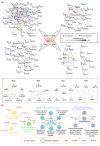
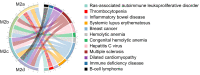
Macrophages are widely distributed immune cells that contribute to tissue homeostasis. Human THP-1 cells have been widely used in various macrophage-associated studies, especially those involving pro-inflammatory M1 and anti-inflammatory M2 phenotypes. However, the molecular characterization of four M2 subtypes (M2a, M2b, M2c, and M2d) derived from THP-1 has not been fully investigated. In this study, we systematically analyzed the protein expression profiles of human THP-1-derived macrophages (M0, M1, M2a, M2b, M2c, and M2d) using quantitative proteomics approaches. The commonly and specially regulated proteins of the four M2 subtypes and their potential biological functions were further investigated. The results showed that M2a and M2b, and M2c and M2d have very similar protein expression profiles. These data could serve as an important resource for studies of macrophages using THP-1 cells, and provide a reference to distinguish different M2 subtypes in macrophage-associated diseases for subsequent clinical research.
Keywords: Biological function; M2 subtype; Macrophage; Proteomics; THP-1 cells.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

