Upregulation of the Long Non-coding RNA LINC01480 Is Associated With Immune Infiltration in Coronary Artery Disease Based on an Immune-Related lncRNA-mRNA Co-expression Network
- PMID: 35557532
- PMCID: PMC9086407
- DOI: 10.3389/fcvm.2022.724262
Upregulation of the Long Non-coding RNA LINC01480 Is Associated With Immune Infiltration in Coronary Artery Disease Based on an Immune-Related lncRNA-mRNA Co-expression Network
Abstract
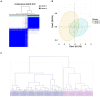
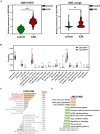
Coronary artery disease (CAD) is considered one of the leading causes of death worldwide. Although dysregulation of long non-coding RNAs (lncRNAs) has been reported to be associated with the initiation and progression of CAD, the knowledge regarding their specific functions as well their physiological/pathological significance in CAD is very limited. In this study, we aimed to systematically analyze immune-related lncRNAs in CAD and explore the relationship between key immune-related lncRNAs and the immune cell infiltration process. Based on differential expression analysis of mRNAs and lncRNAs, an immune-related lncRNA-mRNA weighted gene co-expression network containing 377 lncRNAs and 119 mRNAs was constructed. LINC01480 and AL359237.1 were identified as the hub immune-related lncRNAs in CAD using the random forest-recursive feature elimination and least absolute shrinkage and selection operator logistic regression. Furthermore, 93 CAD samples were divided into two subgroups according to the expression values of LINC01480 and AL359237.1 by consensus clustering analysis. By performing gene set enrichment analysis, we found that cluster 2 enriched more cardiovascular risk pathways than cluster 1. The immune cell infiltration analysis of ischemic cardiomyopathy (ICM; an advanced stage of CAD) samples revealed that the proportion of macrophage M2 was upregulated in the LINC01480 highly expressed samples, thus suggesting that LINC01480 plays a protective role in the progression of ICM. Based on the findings of this study, lncRNA LINC01480 may be used as a novel biomarker and therapeutic target for CAD.
Keywords: atherosclerosis; coronary artery disease; immune molecule; ischemic cardiomyopathy; long noncoding RNA.
Copyright © 2022 Xiong, Xiao, Wu, Liu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

