Persistence of Unintegrated HIV DNA Associates With Ongoing NK Cell Activation and CD34+DNAM-1brightCXCR4+ Precursor Turnover in Vertically Infected Patients Despite Successful Antiretroviral Treatment
- PMID: 35558085
- PMCID: PMC9088003
- DOI: 10.3389/fimmu.2022.847816
Persistence of Unintegrated HIV DNA Associates With Ongoing NK Cell Activation and CD34+DNAM-1brightCXCR4+ Precursor Turnover in Vertically Infected Patients Despite Successful Antiretroviral Treatment
Abstract
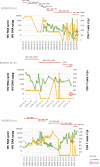
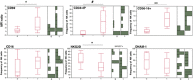
The quantification of proviral DNA is raising interest in view of clinical management and functional HIV eradication. Measures of all unintegrated HIV DNA (uDNA) forms in infected reservoir cells provides information on recent replication events that is not found from other proviral DNA assays. To evaluate its actual relevance in a cohort of perinatally-infected adult HIV patients (PHIV), we studied how peripheral blood mononuclear cell uDNA levels correlated with total HIV DNA (tDNA) and with overall replication or innate immune control parameters including NK cell activation/exhaustion and lymphoid turnover. Twenty-two PHIV were included, with successfully controlled HIV (HIV RNA <50 copies/mL) on combined antiretroviral therapy for mean of 8.7 ± 3.9 years. uDNA accounted for 16 [5.2-83.5] copies/µg and was strongly correlated with tDNA (ρ=0.700, p=0.001). Flow cytometric analysis of peripheral NK cells showed that CD69 expression was directly correlated uDNA (p=0.0412), but not with tDNA. Interestingly, CD56-CD16+NK cells which include newly described inflammatory precursors and terminally differentiated cells were directly correlated with uDNA levels (p<0.001), but not with tDNA, and an inverse association was observed between the proportion of NKG2D+ NK cells and uDNA (ρ=-0.548, p=0.015). In addition, CD34+DNAM-1brightCXCR4+ inflammatory precursor frequency correlated directly with uDNA levels (ρ=0.579, p=0.0075). The frequencies of CD56-CD16+ and CD34+DNAM-1brightCXCR4+ cells maintained association with uDNA levels in a multivariable analysis (p=0.045 and p=0.168, respectively). Thus, control of HIV-1 reservoir in aviremic patients on ART is an active process associated with continuous NK cell intervention and turnover, even after many years of treatment. Quantification of linear and circular uDNA provides relevant information on the requirement for ongoing innate immune control in addition to ART, on recent replication history and may help stratify patients for functional HIV eradication protocols with targeted options.
Keywords: ART; CD34; HIV; inflammatory precursor; natural killer; reservoir; residual replication; unintegrated HIV-DNA.
Copyright © 2022 Taramasso, Bozzano, Casabianca, Orlandi, Bovis, Mora, Giacomini, Moretta, Magnani, Di Biagio and De Maria.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

