Virulence and Host Range of Fungi Associated With the Invasive Plant Ageratina adenophora
- PMID: 35558123
- PMCID: PMC9087049
- DOI: 10.3389/fmicb.2022.857796
Virulence and Host Range of Fungi Associated With the Invasive Plant Ageratina adenophora
Abstract
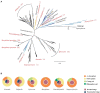
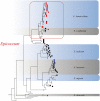
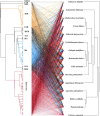
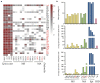
To determine whether disease-mediated invasion of exotic plants can occur and whether this increases the risk of disease transmission in local ecosystems, it is necessary to characterize the species composition and host range of pathogens accumulated in invasive plants. In this study, we found that Didymellaceae, a family containing economically important plant fungal pathogens, is commonly associated with the invasive plant Ageratina adenophora. Accordingly, we characterized its phylogenetic position through multi-locus phylogenetic analysis, as well as its environmental distribution, virulence, and host range. The results indicated that 213 fungal collections were from 11 genera in Didymellaceae, ten of which are known, and one is potentially new. Didymella, Epicoccum, Remotididymella, and Mesophoma were the dominant genera, accounting for 93% of total isolates. The virulence and host ranges of these fungi were related to their phylogenetic relationship. Boeremia exigua, Epicoccum latusicollum, and E. sorghinum were found to be strongly virulent toward all tested native plants as well as toward A. adenophora; M. speciosa and M. ageratinae were weakly virulent toward native plants but strongly virulent toward A. adenophora, thus displaying a narrow host range. Co-evolution analysis showed no strong phylogenetical signal between Didymellaceae and host plants. Isolates S188 and Y122 (belonging to M. speciosa and M. ageratinae, respectively) showed strong virulence toward A. adenophora relative to native plants, highlighting their potential as biocontrol agents for A. adenophora invasion. This study provides new insights into the understanding of the long-term ecological consequences of disease transmission driven by plant invasion.
Keywords: Didymellaceae; biocontrol agent; disease transmission; pathogen accumulation; plant–pathogen co-evolution.
Copyright © 2022 Chen, Yang, Li and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Remotididymella ageratinae sp. nov. and Remotididymella anemophila sp. nov., two novel species isolated from the invasive weed Ageratina adenophora in PR China.Int J Syst Evol Microbiol. 2021 Jan;71(1). doi: 10.1099/ijsem.0.004572. Epub 2020 Nov 18. Int J Syst Evol Microbiol. 2021. PMID: 33206031
-
Diversity and pathogenicity of Alternaria species associated with the invasive plant Ageratina adenophora and local plants.PeerJ. 2022 Feb 28;10:e13012. doi: 10.7717/peerj.13012. eCollection 2022. PeerJ. 2022. PMID: 35251785 Free PMC article.
-
Quantifying the sharing of foliar fungal pathogens by the invasive plant Ageratina adenophora and its neighbours.New Phytol. 2020 Sep;227(5):1493-1504. doi: 10.1111/nph.16624. Epub 2020 May 20. New Phytol. 2020. PMID: 32343409
-
Invasive mechanism and control strategy of Ageratina adenophora (Sprengel).Sci China Life Sci. 2010 Nov;53(11):1291-8. doi: 10.1007/s11427-010-4080-7. Epub 2010 Nov 3. Sci China Life Sci. 2010. PMID: 21046320 Review.
-
An Overview: The Toxicity of Ageratina adenophora on Animals and Its Possible Interventions.Int J Mol Sci. 2021 Oct 27;22(21):11581. doi: 10.3390/ijms222111581. Int J Mol Sci. 2021. PMID: 34769012 Free PMC article. Review.
Cited by
-
Distinct effects of phyllosphere and rhizosphere microbes on invader Ageratina adenophora during its early life stages.Elife. 2024 Jun 19;13:RP95502. doi: 10.7554/eLife.95502. Elife. 2024. PMID: 38896455 Free PMC article.
-
Role of the Foliar Endophyte Colletotrichum in the Resistance of Invasive Ageratina adenophora to Disease and Abiotic Stress.Microorganisms. 2024 Dec 12;12(12):2565. doi: 10.3390/microorganisms12122565. Microorganisms. 2024. PMID: 39770768 Free PMC article.
-
Species evolution: cryptic species and phenotypic noise with a particular focus on fungal systematics.Front Cell Infect Microbiol. 2025 Feb 4;15:1497085. doi: 10.3389/fcimb.2025.1497085. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 39967791 Free PMC article. Review.
References
-
- Beckstead J., Meyer S. E., Connolly B. M., Huck M. B., Street L. E. (2010). Cheatgrass facilitates spillover of a seed bank pathogen onto native grass species. J. Ecol. 98, 168–177. doi: 10.1111/j.1365-2745.2009.01599.x - DOI
-
- Buccellato L., Fisher J. T., Witkowski E. T. F., Byrne M. J. (2021). The effects of a stem gall fly and a leaf pathogen on the reproductive output of Crofton weed, Ageratina adenophora (Asteraceae), in greenhouse and field trials. Biol. Control 152:104453. doi: 10.1016/j.biocontrol.2020.104453 - DOI
LinkOut - more resources
Full Text Sources

