The Interaction of Influenza A NS1 and Cellular TRBP Protein Modulates the Function of RNA Interference Machinery
- PMID: 35558132
- PMCID: PMC9087287
- DOI: 10.3389/fmicb.2022.859420
The Interaction of Influenza A NS1 and Cellular TRBP Protein Modulates the Function of RNA Interference Machinery
Abstract
Influenza A virus (IAV), one of the most prevalent respiratory diseases, causes pandemics around the world. The multifunctional non-structural protein 1 (NS1) of IAV is a viral antagonist that suppresses host antiviral response. However, the mechanism by which NS1 modulates the RNA interference (RNAi) pathway remains unclear. Here, we identified interactions between NS1 proteins of Influenza A/PR8/34 (H1N1; IAV-PR8) and Influenza A/WSN/1/33 (H1N1; IAV-WSN) and Dicer's cofactor TAR-RNA binding protein (TRBP). We found that the N-terminal RNA binding domain (RBD) of NS1 and the first two domains of TRBP protein mediated this interaction. Furthermore, two amino acid residues (Arg at position 38 and Lys at position 41) in NS1 were essential for the interaction. We generated TRBP knockout cells and found that NS1 instead of NS1 mutants (two-point mutations within NS1, R38A/K41A) inhibited the process of microRNA (miRNA) maturation by binding with TRBP. PR8-infected cells showed masking of short hairpin RNA (shRNA)-mediated RNAi, which was not observed after mutant virus-containing NS1 mutation (R38A/K41A, termed PR8/3841) infection. Moreover, abundant viral small interfering RNAs (vsiRNAs) were detected in vitro and in vivo upon PR8/3841 infection. We identify, for the first time, the interaction between NS1 and TRBP that affects host RNAi machinery.
Keywords: RNA interference machinery; TRBP; antiviral RNAi response; influenza A virus; non-structural protein 1 of IAV.
Copyright © 2022 Wang, Wang, Xu, Li, Wang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





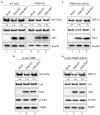


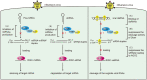
Similar articles
-
Altering Intracellular Localization of the RNA Interference Factors by Influenza A Virus Non-structural Protein 1.Front Microbiol. 2020 Nov 12;11:590904. doi: 10.3389/fmicb.2020.590904. eCollection 2020. Front Microbiol. 2020. PMID: 33281788 Free PMC article.
-
A Promising IFN-Deficient System to Manufacture IFN-Sensitive Influenza Vaccine Virus.Front Cell Infect Microbiol. 2018 May 1;8:127. doi: 10.3389/fcimb.2018.00127. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29765910 Free PMC article.
-
Phosphorylation of Influenza A Virus NS1 at Serine 205 Mediates Its Viral Polymerase-Enhancing Function.J Virol. 2021 Feb 24;95(6):e02369-20. doi: 10.1128/JVI.02369-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33408177 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Discovery of Small Molecule Influenza Virus NS1 Antagonist.2012 Apr 13 [updated 2013 Sep 3]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. 2012 Apr 13 [updated 2013 Sep 3]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. PMID: 24260784 Free Books & Documents. Review.
Cited by
-
The subgenomic flaviviral RNA suppresses RNA interference through competing with siRNAs for binding RISC components.J Virol. 2024 Feb 20;98(2):e0195423. doi: 10.1128/jvi.01954-23. Epub 2024 Jan 30. J Virol. 2024. PMID: 38289102 Free PMC article.
-
Deficiency of IFNAR1 Increases the Production of Influenza Vaccine Viruses in MDCK Cells.Viruses. 2025 Aug 8;17(8):1097. doi: 10.3390/v17081097. Viruses. 2025. PMID: 40872812 Free PMC article.
-
Impact of the Transboundary Interference Inhibitor on RNAi and the Baculovirus Expression System in Insect Cells.Insects. 2024 May 21;15(6):375. doi: 10.3390/insects15060375. Insects. 2024. PMID: 38921090 Free PMC article.
-
Research progress on the nonstructural protein 1 (NS1) of influenza a virus.Virulence. 2024 Dec;15(1):2359470. doi: 10.1080/21505594.2024.2359470. Epub 2024 Jun 25. Virulence. 2024. PMID: 38918890 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

