NPM1 and DNMT3A mutations are associated with distinct blast immunophenotype in acute myeloid leukemia
- PMID: 35558161
- PMCID: PMC9090295
- DOI: 10.1080/2162402X.2022.2073050
NPM1 and DNMT3A mutations are associated with distinct blast immunophenotype in acute myeloid leukemia
Abstract
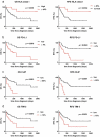
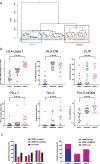
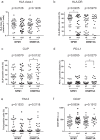
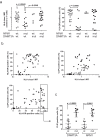
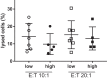
The immune system is important for elimination of residual leukemic cells during acute myeloid leukemia (AML) therapy. Anti-leukemia immune response can be inhibited by various mechanisms leading to immune evasion and disease relapse. Selected markers of immune escape were analyzed on AML cells from leukapheresis at diagnosis (N = 53). Hierarchical clustering of AML immunophenotypes yielded distinct genetic clusters. In the absence of DNMT3A mutation, NPM1 mutation was associated with decreased HLA expression and low levels of other markers (CLIP, PD-L1, TIM-3). Analysis of an independent cohort confirmed decreased levels of HLA transcripts in patients with NPM1 mutation. Samples with combined NPM1 and DNMT3A mutations had high CLIP surface amount suggesting reduced antigen presentation. TIM-3 transcript correlated not only with TIM-3 surface protein but also with CLIP and PD-L1. In our cohort, high levels of TIM-3/PD-L1/CLIP were associated with lower survival. Our results suggest that AML genotype is related to blast immunophenotype, and that high TIM-3 transcript levels in AML blasts could be a marker of immune escape. Cellular pathways regulating resistance to the immune system might contribute to the predicted response to standard therapy of patients in specific AML subgroups and should be targeted to improve AML treatment.
Keywords: AML; DNMT3A; NPM1; TIM-3; immunophenotype.
© 2022 The Author(s). Published with license by Taylor & Francis Group, LLC.
Conflict of interest statement
The authors report there are no competing interests to declare.
Figures

References
-
- Buggins AG, Milojkovic D, Arno MJ, Lea NC, Mufti GJ, Thomas NS, Hirst WJR.. Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J Immunol. 2001;167(10):6021–14. doi: 10.4049/jimmunol.167.10.6021. - DOI - PubMed
-
- Knaus HA, Berglund S, Hackl H, Blackford AL, Zeidner JF, Montiel-Esparza R, Mukhopadhyay R, Vanura K, Blazar BR, Karp JE, et al. Signatures of CD8+ T cell dysfunction in AML patients and their reversibility with response to chemotherapy. JCI Insight. 2018;3(21):e120974. doi: 10.1172/jci.insight.120974. - DOI - PMC - PubMed
-
- Le Dieu R, Taussig DC, Ramsay AG, Mitter R, Miraki-Moud F, Fatah R, Lee AM, Lister TA, Gribben JG.. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114(18):3909–3916. doi: 10.1182/blood-2009-02-206946. - DOI - PMC - PubMed
-
- Lamble AJ, Kosaka Y, Laderas T, Maffit A, Kaempf A, Brady LK, Wang W, Long N, Saultz JN, Mori M, et al. Reversible suppression of T cell function in the bone marrow microenvironment of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2020;117(25):14331–14341. doi: 10.1073/pnas.1916206117. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
