Gut microbiota as important modulator of metabolism in health and disease
- PMID: 35558413
- PMCID: PMC9092240
- DOI: 10.1039/c8ra08094a
Gut microbiota as important modulator of metabolism in health and disease
Abstract
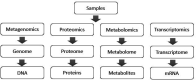
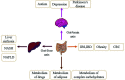
The human gastrointestinal tract colonizes a large number of microbial microflora, forms a host-microbiota co-metabolism structure with the host to participate in various metabolic processes in the human body, and plays a major role in the host immune response. In addition, the dysbiosis of intestinal microbial homeostasis is closely related to many diseases. Thus, an in-depth understanding of the relationship between them is of importance for disease pathogenesis, prevention and treatment. The combined use of metagenomics, transcriptomics, proteomics and metabolomics techniques for the analysis of gut microbiota can reveal the relationship between microbiota and the host in many ways, which has become a hot topic of analysis in recent years. This review describes the mechanism of co-metabolites in host health, including short-chain fatty acids (SCFA) and bile acid metabolism. The metabolic role of gut microbiota in obesity, liver diseases, gastrointestinal diseases and other diseases is also summarized, and the research methods for multi-omics combined application on gut microbiota are summarized. According to the studies of the interaction mechanism between gut microbiota and the host, we have a better understanding of the use of intestinal microflora in the treatment of related diseases. It is hoped that the gut microbiota can be utilized to maintain human health, providing a reference for future research.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
Similar articles
-
Gut microbiota and host metabolism in liver cirrhosis.World J Gastroenterol. 2015 Nov 7;21(41):11597-608. doi: 10.3748/wjg.v21.i41.11597. World J Gastroenterol. 2015. PMID: 26556989 Free PMC article. Review.
-
Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex.J Autoimmun. 2018 Aug;92:12-34. doi: 10.1016/j.jaut.2018.05.008. Epub 2018 Jun 1. J Autoimmun. 2018. PMID: 29861127 Review.
-
The impact of metabolites derived from the gut microbiota on immune regulation and diseases.Int Immunol. 2020 Sep 30;32(10):629-636. doi: 10.1093/intimm/dxaa041. Int Immunol. 2020. PMID: 32564086 Review.
-
Farnesoid X Receptor Signaling Shapes the Gut Microbiota and Controls Hepatic Lipid Metabolism.mSystems. 2016 Oct 11;1(5):e00070-16. doi: 10.1128/mSystems.00070-16. eCollection 2016 Sep-Oct. mSystems. 2016. PMID: 27822554 Free PMC article.
-
[Gut microbiota and immune crosstalk in metabolic disease].Biol Aujourdhui. 2017;211(1):1-18. doi: 10.1051/jbio/2017008. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682223 Review. French.
Cited by
-
Dietary Supplementation with Sea Buckthorn Berry Puree Alters Plasma Metabolomic Profile and Gut Microbiota Composition in Hypercholesterolemia Population.Foods. 2022 Aug 17;11(16):2481. doi: 10.3390/foods11162481. Foods. 2022. PMID: 36010480 Free PMC article.
-
Lignans and Gut Microbiota: An Interplay Revealing Potential Health Implications.Molecules. 2020 Dec 3;25(23):5709. doi: 10.3390/molecules25235709. Molecules. 2020. PMID: 33287261 Free PMC article. Review.
-
Chemical interplay between gut microbiota and epigenetics: Implications in circadian biology.Cell Chem Biol. 2025 Jan 16;32(1):61-82. doi: 10.1016/j.chembiol.2024.04.016. Epub 2024 May 21. Cell Chem Biol. 2025. PMID: 38776923 Review.
-
Determinants of overweight and obesity and other cardiometabolic risks in adolescents: a Spanish longitudinal birth study.Pediatr Res. 2025 Jul 18. doi: 10.1038/s41390-025-04273-w. Online ahead of print. Pediatr Res. 2025. PMID: 40681697
-
An integrated metabolomics and 16S rRNA gene sequencing approach exploring the molecular pathways and potential targets behind the effects of Radix Scrophulariae.RSC Adv. 2019 Oct 17;9(57):33354-33367. doi: 10.1039/c9ra03912k. eCollection 2019 Oct 15. RSC Adv. 2019. PMID: 35529111 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

