Polydopamine nanoparticles kill cancer cells
- PMID: 35558470
- PMCID: PMC9088449
- DOI: 10.1039/c8ra05586f
Polydopamine nanoparticles kill cancer cells
Abstract
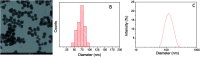
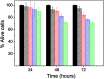
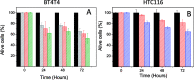
Polydopamine (PD) is a synthetic melanin analogue of growing importance in the field of biomedicine, especially with respect to cancer research, due, in part, to its biocompatibility. But little is known about the cytotoxic effects of PD on cancer cell lines. PD is a UV-vis absorbing material whose absorbance overlaps with that of formazan salts, which are used to assess cell viability in MTT assays. In this study, a protocol has been established to eliminate the contributing absorbance of PD at 550 nm, and has been applied to characterize the cytotoxicity of PD nanoparticles in both healthy and breast cancer cell lines. Once the protocol is applied, it was found that PD is per se an antineoplastic system, meaning it selectively kills cancer cells, especially those of breast cancer, but it has no toxic effect on healthy cells. The mechanism of action could be related to the production of ROS and the alteration of iron homeostasis in lysosomes. To the best of our knowledge there are only a few examples of nanoparticle systems devoid of drugs that selectively kill cancer cells.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interests.
Figures




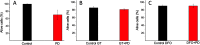
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

