Local Ion Densities can Influence Transition Paths of Molecular Binding
- PMID: 35558558
- PMCID: PMC9086317
- DOI: 10.3389/fmolb.2022.858316
Local Ion Densities can Influence Transition Paths of Molecular Binding
Abstract
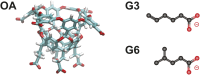
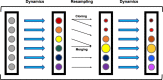
Improper reaction coordinates can pose significant problems for path-based binding free energy calculations. Particularly, omission of long timescale motions can lead to over-estimation of the energetic barriers between the bound and unbound states. Many methods exist to construct the optimal reaction coordinate using a pre-defined basis set of features. Although simulations are typically conducted in explicit solvent, the solvent atoms are often excluded by these feature sets-resulting in little being known about their role in reaction coordinates, and ultimately, their role in determining (un)binding rates and free energies. In this work, analysis is done on an extensive set of host-guest unbinding trajectories, working to characterize differences between high and low probability unbinding trajectories with a focus on solvent-based features, including host-ion interactions, guest-ion interactions and location-dependent ion densities. We find that differences in ion densities as well as guest-ion interactions strongly correlate with differences in the probabilities of reactive paths that are used to determine free energies of (un)binding and play a significant role in the unbinding process.
Keywords: SAMPL system; binding affinity; free energy; ligand unbinding; mechanisms; molecular dynamics; weighted ensemble.
Copyright © 2022 Roussey and Dickson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Quality over quantity: Sampling high probability rare events with the weighted ensemble algorithm.J Comput Chem. 2023 Mar 30;44(8):935-947. doi: 10.1002/jcc.27054. Epub 2022 Dec 13. J Comput Chem. 2023. PMID: 36510846 Free PMC article.
-
Combined Free-Energy Calculation and Machine Learning Methods for Understanding Ligand Unbinding Kinetics.J Chem Theory Comput. 2022 Apr 12;18(4):2543-2555. doi: 10.1021/acs.jctc.1c00924. Epub 2022 Feb 23. J Chem Theory Comput. 2022. PMID: 35195418 Free PMC article.
-
Variational implicit-solvent predictions of the dry-wet transition pathways for ligand-receptor binding and unbinding kinetics.Proc Natl Acad Sci U S A. 2019 Jul 23;116(30):14989-14994. doi: 10.1073/pnas.1902719116. Epub 2019 Jul 3. Proc Natl Acad Sci U S A. 2019. PMID: 31270236 Free PMC article.
-
Overview of the SAMPL5 host-guest challenge: Are we doing better?J Comput Aided Mol Des. 2017 Jan;31(1):1-19. doi: 10.1007/s10822-016-9974-4. Epub 2016 Sep 22. J Comput Aided Mol Des. 2017. PMID: 27658802 Free PMC article. Review.
-
The importance of ensemble averaging in enzyme kinetics.Acc Chem Res. 2015 Feb 17;48(2):431-8. doi: 10.1021/ar500319e. Epub 2014 Dec 24. Acc Chem Res. 2015. PMID: 25539028 Free PMC article. Review.
Cited by
-
Quality over quantity: Sampling high probability rare events with the weighted ensemble algorithm.J Comput Chem. 2023 Mar 30;44(8):935-947. doi: 10.1002/jcc.27054. Epub 2022 Dec 13. J Comput Chem. 2023. PMID: 36510846 Free PMC article.
-
Water regulates the residence time of Benzamidine in Trypsin.Nat Commun. 2022 Sep 16;13(1):5438. doi: 10.1038/s41467-022-33104-3. Nat Commun. 2022. PMID: 36114175 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

