Identification of a novel ene reductase from Pichia angusta with potential application in (R)-levodione production
- PMID: 35558851
- PMCID: PMC9088392
- DOI: 10.1039/d2ra01716d
Identification of a novel ene reductase from Pichia angusta with potential application in (R)-levodione production
Abstract
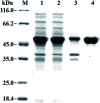
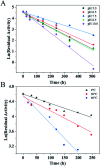
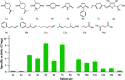
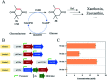
Asymmetric reduction of electronically activated alkenes by ene reductases (ERs) is an attractive approach for the production of enantiopure chiral products. Herein, a novel FMN-binding ene reductase (PaER) from Pichia angusta was heterologously expressed in Escherichia coli BL21(DE3), and the recombinant enzyme was characterized for its biocatalytic properties. PaER displayed optimal activity at 40 °C and pH 7.5, respectively. The purified enzyme was quite stable below 30 °C over a broad pH range of 5.0-10.0. PaER was identified to have a good ability to reduce the C[double bond, length as m-dash]C bond of various α,β-unsaturated compounds in the presence of NADPH. In addition, PaER exhibited a high reduction rate (k cat = 3.57 s-1) and an excellent stereoselectivity (>99%) for ketoisophorone. Engineered E. coli cells harboring PaER and glucose dehydrogenase (for cofactor regeneration) were employed as biocatalysts for the asymmetric reduction of ketoisophorone. As a result, up to 1000 mM ketoisophorone was completely and enantioselectively converted to (R)-levodione with a >99% ee value in a space-time yield of 460.7 g L-1 d-1. This study provides a great potential biocatalyst for practical synthesis of (R)-levodione.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
An ene reductase from Clavispora lusitaniae for asymmetric reduction of activated alkenes.Enzyme Microb Technol. 2014 Mar 5;56:40-5. doi: 10.1016/j.enzmictec.2013.12.016. Epub 2014 Jan 8. Enzyme Microb Technol. 2014. PMID: 24564901
-
Identification and characterization of an ene-reductase from Corynebacterium casei.Int J Biol Macromol. 2024 Apr;264(Pt 1):130427. doi: 10.1016/j.ijbiomac.2024.130427. Epub 2024 Feb 29. Int J Biol Macromol. 2024. PMID: 38428763
-
Comparative characterization of novel ene-reductases from cyanobacteria.Biotechnol Bioeng. 2013 May;110(5):1293-301. doi: 10.1002/bit.24817. Epub 2013 Jan 21. Biotechnol Bioeng. 2013. PMID: 23280373
-
Application of protein engineering to ene-reductase for the synthesis of chiral compounds through asymmetric reaction.Crit Rev Biotechnol. 2025 May;45(3):665-682. doi: 10.1080/07388551.2024.2382957. Epub 2024 Aug 12. Crit Rev Biotechnol. 2025. PMID: 39134447 Review.
-
Ene-Reductase: A Multifaceted Biocatalyst in Organic Synthesis.Chemistry. 2022 Apr 12;28(21):e202103949. doi: 10.1002/chem.202103949. Epub 2022 Mar 3. Chemistry. 2022. PMID: 35133702 Review.
Cited by
-
Applications of Ene-Reductases in the Synthesis of Flavors and Fragrances.J Agric Food Chem. 2024 Aug 21;72(33):18305-18320. doi: 10.1021/acs.jafc.4c02897. Epub 2024 Jul 5. J Agric Food Chem. 2024. PMID: 38966982 Free PMC article. Review.
-
Recombinant Production and Characterization of Six Ene-reductases from Penicillium steckii.Chembiochem. 2025 Apr 1;26(7):e202401007. doi: 10.1002/cbic.202401007. Epub 2025 Mar 24. Chembiochem. 2025. PMID: 40072226 Free PMC article.
References
-
- Zuliani A. Cova C. M. Manno R. Sebastian V. Romero A. A. Luque R. Green Chem. 2020;22:379–387. doi: 10.1039/C9GC03299A. - DOI
-
- Scholtissek A. Tischler D. Westphal A. van Berkel W. Paul C. Catalysts. 2017;7:130. doi: 10.3390/catal7050130. - DOI
-
- Zheng L. Lin J. Zhang B. Kuang Y. Wei D. Bioresour. Bioprocess. 2018;5:9. doi: 10.1186/s40643-018-0192-x. - DOI
-
- Hall M. Hauer B. Stuermer R. Kroutil W. Faber K. Tetrahedron: Asymmetry. 2006;17:3058–3062. doi: 10.1016/j.tetasy.2006.11.018. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

